7A0Q
 
 | |
7A0T
 
 | |
5K3W
 
 | | Structural characterisation of fold IV-transaminase, CpuTA1, from Curtobacterium pusillum | | Descriptor: | 3-AMINOBENZOIC ACID, CpuTA1, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Pavkov-Keller, T, Diepold, M, Steiner, K, Gruber, K. | | Deposit date: | 2016-05-20 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Discovery and structural characterisation of new fold type IV-transaminases exemplify the diversity of this enzyme fold.
Sci Rep, 6, 2016
|
|
5M3K
 
 | | A multi-component acyltransferase PhlABC from Pseudomonas protegens | | Descriptor: | 2,4-diacetylphloroglucinol biosynthesis protein PhlB, 2,4-diacetylphloroglucinol biosynthesis protein PhlC, PhlA, ... | | Authors: | Pavkov-Keller, T, Schmidt, N.G, Kroutil, W, Gruber, K. | | Deposit date: | 2016-10-15 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structure and Catalytic Mechanism of a Bacterial Friedel-Crafts Acylase.
Chembiochem, 20, 2019
|
|
5M58
 
 | |
5MG5
 
 | | A multi-component acyltransferase PhlABC from Pseudomonas protegens soaked with the monoacetylphloroglucinol (MAPG) | | Descriptor: | 2,4-diacetylphloroglucinol biosynthesis protein, 2,4-diacetylphloroglucinol biosynthesis protein PhlC, Hydroxymethylglutaryl-CoA synthase, ... | | Authors: | Pavkov-Keller, T, Schmidt, N.G, Kroutil, W, Gruber, K. | | Deposit date: | 2016-11-20 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.44 Å) | | Cite: | Structure and Catalytic Mechanism of a Bacterial Friedel-Crafts Acylase.
Chembiochem, 20, 2019
|
|
4UIC
 
 | |
4UID
 
 | |
4UIE
 
 | | Crystal structure of the S-layer protein SbsC, domains 7, 8 and 9 | | Descriptor: | CALCIUM ION, OSMIUM ION, SURFACE LAYER PROTEIN | | Authors: | Pavkov-Keller, T, Dordic, A, Egelseer, E.M, Sleytr, U.B, Keller, W. | | Deposit date: | 2015-03-27 | | Release date: | 2016-04-13 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of the S-Layer Protein Sbsc
To be Published
|
|
4BCT
 
 | | Crystal structure of kiwi-fruit allergen Act d 2 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, THAUMATIN-LIKE PROTEIN | | Authors: | Pavkov-Keller, T, Bublin, M, Jankovic, M, Breiteneder, H, Keller, W. | | Deposit date: | 2012-10-03 | | Release date: | 2013-10-16 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Crystal Structure of Kiwi-Fruit Allergen Act D 2
To be Published
|
|
5EB4
 
 | | The crystal structure of almond HNL, PaHNL5 V317A, expressed in Aspergillus niger | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Pavkov-Keller, T, Steinkellner, G, Gruber, K. | | Deposit date: | 2015-10-17 | | Release date: | 2016-04-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of almond hydroxynitrile lyase isoenzyme 5 provide a rationale for the lack of oxidoreductase activity in flavin dependent HNLs.
J.Biotechnol., 235, 2016
|
|
5EB5
 
 | | The crystal structure of almond HNL, PaHNL5 V317A, in complex with benzyl alcohol | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Pavkov-Keller, T, Steinkellner, G, Gruber, K. | | Deposit date: | 2015-10-17 | | Release date: | 2016-04-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of almond hydroxynitrile lyase isoenzyme 5 provide a rationale for the lack of oxidoreductase activity in flavin dependent HNLs.
J.Biotechnol., 235, 2016
|
|
4V39
 
 | | Apo-structure of alpha2,3-sialyltransferase variant 2 from Pasteurella dagmatis | | Descriptor: | SIALYLTRANSFERASE | | Authors: | Pavkov-Keller, T, Schmoelzer, K, Czabany, T, Luley-Goedl, C, Ribitsch, D, Schwab, H, Nidetzky, B, Gruber, K. | | Deposit date: | 2014-10-17 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Complete Switch from Alpha2,3- to Alpha2,6-Regioselectivity in Pasteurella Dagmatis Beta-D-Galactoside Sialyltransferase by Active-Site Redesign
Chem.Commun.(Camb.), 51, 2015
|
|
4V3C
 
 | | The structure of alpha2,3-sialyltransferase variant 2 from Pasteurella dagmatis in complex with the donor product CMP | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, SIALYLTRANSFERASE | | Authors: | Pavkov-Keller, T, Schmoelzer, K, Czabany, T, Luley-Goedl, C, Ribitsch, D, Schwab, H, Nidetzky, B, Gruber, K. | | Deposit date: | 2014-10-17 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Complete Switch from Alpha2,3- to Alpha2,6-Regioselectivity in Pasteurella Dagmatis Beta-D-Galactoside Sialyltransferase by Active-Site Redesign
Chem.Commun.(Camb.), 51, 2015
|
|
4V3B
 
 | | The structure of alpha2,3-sialyltransferase variant 1 from Pasteurella dagmatis in complex with the donor product CMP | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, SIALYLTRANSFERASE | | Authors: | Pavkov-Keller, T, Schmoelzer, K, Czabany, T, Luley-Goedl, C, Ribitsch, D, Schwab, H, Nidetzky, B, Gruber, K. | | Deposit date: | 2014-10-17 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Complete Switch from Alpha2,3- to Alpha2,6-Regioselectivity in Pasteurella Dagmatis Beta-D-Galactoside Sialyltransferase by Active-Site Redesign
Chem.Commun.(Camb.), 51, 2015
|
|
4V38
 
 | | Apo-structure of alpha2,3-sialyltransferase variant 1 from Pasteurella dagmatis | | Descriptor: | SIALYLTRANSFERASE | | Authors: | Pavkov-Keller, T, Schmoelzer, K, Czabany, T, Luley-Goedl, C, Ribitsch, D, Schwab, H, Nidetzky, B, Gruber, K. | | Deposit date: | 2014-10-17 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Complete Switch from Alpha2,3- to Alpha2,6-Regioselectivity in Pasteurella Dagmatis Beta-D-Galactoside Sialyltransferase by Active-Site Redesign
Chem.Commun.(Camb.), 51, 2015
|
|
4V2U
 
 | | Apo-structure of alpha2,3-sialyltransferase from Pasteurella dagmatis | | Descriptor: | SIALYLTRANSFERASE | | Authors: | Pavkov-Keller, T, Schmoelzer, K, Czabany, T, Luley-Goedl, C, Ribitsch, D, Schwab, H, Nidetzky, B, Gruber, K. | | Deposit date: | 2014-10-15 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Complete Switch from Alpha2,3- to Alpha2,6-Regioselectivity in Pasteurella Dagmatis Beta-D-Galactoside Sialyltransferase by Active-Site Redesign
Chem.Commun.(Camb.), 51, 2015
|
|
4UIR
 
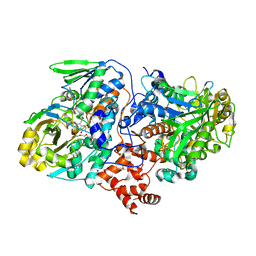 | | Structure of oleate hydratase from Elizabethkingia meningoseptica | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, HEXAETHYLENE GLYCOL, OLEATE HYDRATASE, ... | | Authors: | Pavkov-Keller, T, Hromic, A, Engleder, M, Emmerstorfer, A, Steinkellner, G, Schrempf, S, Wriessnegger, T, Leitner, E, Strohmeier, G.A, Kaluzna, I, Mink, D, Schuermann, M, Wallner, S, Macheroux, P, Pichler, H, Gruber, K. | | Deposit date: | 2015-04-02 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-Based Mechanism of Oleate Hydratase from Elizabethkingia Meningoseptica.
Chembiochem, 16, 2015
|
|
4UXA
 
 | | Improved variant of (R)-selective manganese-dependent hydroxynitrile lyase from bacteria | | Descriptor: | CUPIN 2 CONSERVED BARREL DOMAIN PROTEIN, MANGANESE (II) ION | | Authors: | Pavkov-Keller, T, Wiedner, R, Kothbauer, B, Gruber-Khadjawi, M, Schwab, H, Steiner, K, Gruber, K. | | Deposit date: | 2014-08-21 | | Release date: | 2015-01-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Improving the Properties of Bacterial R-Selective Hydroxynitrile Lyases for Industrial Applications
Chemcatchem, 2015
|
|
4UJ7
 
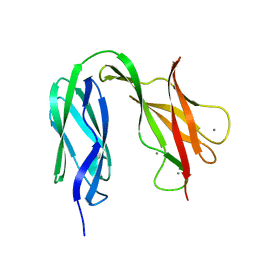 | | Structure of the S-layer protein SbsC, domains 5-6 | | Descriptor: | CALCIUM ION, SURFACE LAYER PROTEIN | | Authors: | Dordic, A, Pavkov-Keller, T, Eder, M, Egelseer, E.M, Davis, K, Mills, D, Sleytr, U.B, Kuehlbrandt, W, Vonck, J, Keller, W. | | Deposit date: | 2015-04-08 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structure of the S-Layer Protein Sbsc, Domains 5-6
To be Published
|
|
4UJ8
 
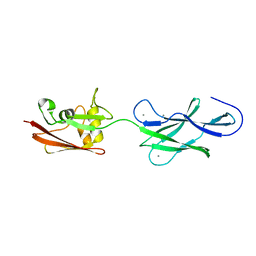 | | Structure of surface layer protein SbsC, domains 6-7 | | Descriptor: | CALCIUM ION, SURFACE LAYER PROTEIN | | Authors: | Dordic, A, Pavkov-Keller, T, Eder, M, Egelseer, E.M, Davis, K, Mills, D, Sleytr, U.B, Kuehlbrandt, W, Vonck, J, Keller, W. | | Deposit date: | 2015-04-08 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Surface Layer Protein Sbsc
To be Published
|
|
4UJ6
 
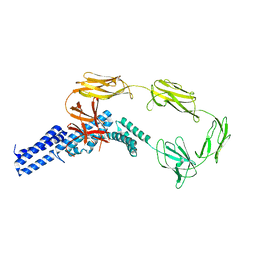 | | Structure of surface layer protein SbsC, domains 1-6 | | Descriptor: | SURFACE LAYER PROTEIN | | Authors: | Dordic, A, Pavkov-Keller, T, Eder, M, Egelseer, E.M, Davis, K, Mills, D, Sleytr, U.B, Kuehlbrandt, W, Vonck, J, Keller, W. | | Deposit date: | 2015-04-08 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of Surface Layer Protein Sbsc, Domains 1-6
To be Published
|
|
6EZD
 
 | |
6GG2
 
 | | The structure of FsqB from Aspergillus fumigatus, a flavoenzyme of the amine oxidase family | | Descriptor: | Amino acid oxidase fmpA, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Pavkov-Keller, T, Lahham, M, Macheroux, P, Gruber, K. | | Deposit date: | 2018-05-02 | | Release date: | 2018-09-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Oxidative cyclization ofN-methyl-dopa by a fungal flavoenzyme of the amine oxidase family.
J. Biol. Chem., 293, 2018
|
|
6GHW
 
 | | Substituting the prolines of 4-oxalocrotonate tautomerase with non-canonical analogue (2S)-3,4-dehydroproline | | Descriptor: | 2-hydroxymuconate tautomerase, CALCIUM ION | | Authors: | Pavkov-Keller, T, Lukesch, M.S, Wiltschi, B, Gruber, K. | | Deposit date: | 2018-05-09 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substituting the catalytic proline of 4-oxalocrotonate tautomerase with non-canonical analogues reveals a finely tuned catalytic system.
Sci Rep, 9, 2019
|
|
