2ES4
 
 | | Crystal structure of the Burkholderia glumae lipase-specific foldase in complex with its cognate lipase | | Descriptor: | CALCIUM ION, IODIDE ION, Lipase, ... | | Authors: | Pauwels, K, Wyns, L, Tommassen, J, Savvides, S.N, Van Gelder, P. | | Deposit date: | 2005-10-25 | | Release date: | 2006-03-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of a membrane-based steric chaperone in complex with its lipase substrate.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2W7Q
 
 | | Structure of Pseudomonas aeruginosa LolA | | Descriptor: | GLYCEROL, OUTER-MEMBRANE LIPOPROTEIN CARRIER PROTEIN | | Authors: | Remans, K, Pauwels, K, van Ulsen, P, Savvides, S, Decanniere, K, Cornelis, P, Tommassen, J, Van Gelder, P. | | Deposit date: | 2008-12-23 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Hydrophobic Surface Patches on Lola of Pseudomonas Aeruginosa are Essential for Lipoprotein Binding.
J.Mol.Biol., 401, 2010
|
|
2KTM
 
 | | Solution NMR structure of H2H3 domain of ovine prion protein (residues 167-234) | | Descriptor: | Major prion protein | | Authors: | Pastore, A, Adrover, M, Pauwels, K, de Chiara, C, Prigent, S, Rezeai, H. | | Deposit date: | 2010-02-04 | | Release date: | 2010-04-07 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Prion fibrillization is mediated by a native structural element that comprises helices H2 and H3.
J.Biol.Chem., 285, 2010
|
|
2N0E
 
 | | NMR structure of Neuromedin C in 40% TFE | | Descriptor: | Neuromedin C (NMC) | | Authors: | Adrover, M, Sanchis, P, Vilanova, B, Pauwels, K, Martorell, G, Perez, J. | | Deposit date: | 2015-03-05 | | Release date: | 2015-10-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Conformational ensembles of neuromedin C reveal a progressive coil-helix transition within a binding-induced folding mechanism.
RSC ADV, 5, 2015
|
|
2N0F
 
 | | NMR structure of Neuromedin C in 60% TFE | | Descriptor: | Neuromedin C (NMC) | | Authors: | Adrover, M, Sanchis, P, Vilanova, B, Pauwels, K, Martorell, G, Perez, J. | | Deposit date: | 2015-03-05 | | Release date: | 2015-10-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Conformational ensembles of neuromedin C reveal a progressive coil-helix transition within a binding-induced folding mechanism.
RSC ADV, 5, 2015
|
|
2N0C
 
 | | NMR structure of Neuromedin C in 10% TFE | | Descriptor: | Neuromedin C (NMC) | | Authors: | Adrover, M, Sanchis, P, Vilanova, B, Pauwels, K, Martorell, G, Perez, J. | | Deposit date: | 2015-03-05 | | Release date: | 2015-10-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Conformational ensembles of neuromedin C reveal a progressive coil-helix transition within a binding-induced folding mechanism.
RSC ADV, 5, 2015
|
|
2N0H
 
 | | NMR structure of Neuromedin C in presence of SDS micelles | | Descriptor: | Neuromedin C (NMC) | | Authors: | Adrover, M, Sanchis, P, Vilanova, B, Pauwels, K, Martorell, G, Perez, J. | | Deposit date: | 2015-03-05 | | Release date: | 2015-10-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Conformational ensembles of neuromedin C reveal a progressive coil-helix transition within a binding-induced folding mechanism.
RSC ADV, 5, 2015
|
|
2N0B
 
 | | NMR structure of Neuromedin C in aqueous solution | | Descriptor: | Neuromedin C (NMC) | | Authors: | Adrover, M, Sanchis, P, Vilanova, B, Pauwels, K, Martorell, G, Perez, J. | | Deposit date: | 2015-03-05 | | Release date: | 2015-10-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Conformational ensembles of neuromedin C reveal a progressive coil-helix transition within a binding-induced folding mechanism.
RSC ADV, 5, 2015
|
|
2N0G
 
 | | NMR structure of Neuromedin C in 90% TFE | | Descriptor: | Neuromedin C (NMC) | | Authors: | Adrover, M, Sanchis, P, Vilanova, B, Pauwels, K, Martorell, G, Perez, J. | | Deposit date: | 2015-03-05 | | Release date: | 2015-10-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Conformational ensembles of neuromedin C reveal a progressive coil-helix transition within a binding-induced folding mechanism.
RSC ADV, 5, 2015
|
|
2N0D
 
 | | NMR structure of Neuromedin C in 25% TFE | | Descriptor: | Neuromedin C (NMC) | | Authors: | Adrover, M, Sanchis, P, Vilanova, B, Pauwels, K, Martorell, G, Perez, J. | | Deposit date: | 2015-03-05 | | Release date: | 2015-10-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Conformational ensembles of neuromedin C reveal a progressive coil-helix transition within a binding-induced folding mechanism.
RSC ADV, 5, 2015
|
|
5J13
 
 | |
5J11
 
 | | Structure of human TSLP in complex with TSLPR and IL-7Ralpha | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokine receptor-like factor 2, Interleukin-7 receptor subunit alpha, ... | | Authors: | Verstraete, K, Savvides, S.N. | | Deposit date: | 2016-03-28 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structure and antagonism of the receptor complex mediated by human TSLP in allergy and asthma.
Nat Commun, 8, 2017
|
|
5D28
 
 | |
5D22
 
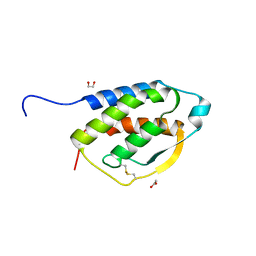 | |
4APT
 
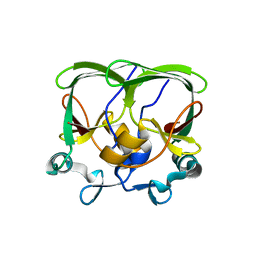 | | The structure of the AXH domain of ataxin-1. | | Descriptor: | ATAXIN-1, SODIUM ION | | Authors: | Rees, M, Chen, Y.W, de Chiara, C, Pastore, A. | | Deposit date: | 2012-04-05 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Self-Assembly and Conformational Heterogeneity of the Axh Domain of Ataxin-1: An Unusual Example of a Chameleon Fold
Biophys.J., 104, 2013
|
|
4AQP
 
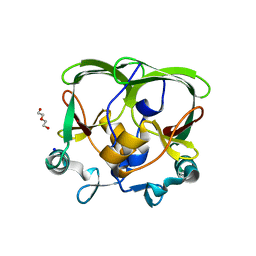 | | The structure of the AXH domain of ataxin-1. | | Descriptor: | ATAXIN-1, DI(HYDROXYETHYL)ETHER, SODIUM ION | | Authors: | Rees, M, Chen, Y.W, de Chiara, C, Pastore, A. | | Deposit date: | 2012-04-19 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.452 Å) | | Cite: | Self-Assembly and Conformational Heterogeneity of the Axh Domain of Ataxin-1: An Unusual Example of a Chameleon Fold
Biophys.J., 104, 2013
|
|
