2Z71
 
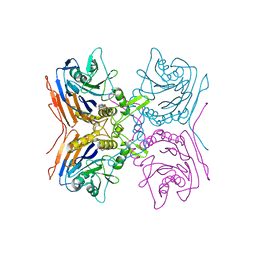 | | Structure of truncated mutant CYS1GLY of penicillin V acylase from bacillus sphaericus co-crystallized with penicillin V | | Descriptor: | (2S,5R,6R)-3,3-DIMETHYL-7-OXO-6-(2-PHENOXYACETAMIDO)-4-THIA-1- AZABICYCLO(3.2.0)HEPTANE-2-CARBOXYLIC ACID, Penicillin acylase | | Authors: | Pathak, M.C, Brannigan, J, Dodson, G.G, Suresh, C.G. | | Deposit date: | 2007-08-10 | | Release date: | 2008-08-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Studies on the catalysis and post translational processing of penicillin V acylase
To be published
|
|
2QUY
 
 | |
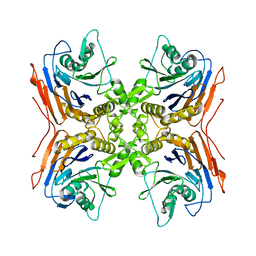
3MJI
 
 | |
4GAH
 
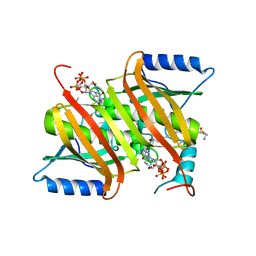 | | Human acyl-CoA thioesterases 4 in complex with undecan-2-one-CoA inhibitor | | Descriptor: | Thioesterase superfamily member 4, [[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(3R)-2,2-dimethyl-3-oxidanyl-4-oxidanylidene-4-[[3-oxidanylidene-3-[2-[(2R)-2-oxidanylundecyl]sulfanylethylamino]propyl]amino]butyl] hydrogen phosphate | | Authors: | Lim, K, Pathak, M.C, Herzberg, O. | | Deposit date: | 2012-07-25 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Correlation of structure and function in the human hotdog-fold enzyme hTHEM4.
Biochemistry, 51, 2012
|
|
4M8Z
 
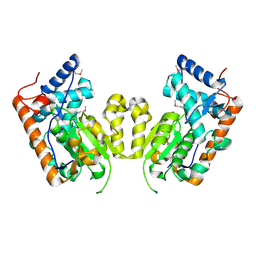 | |
4IS8
 
 | |
4DOR
 
 | |
4DOS
 
 | | Human Nuclear Receptor Liver Receptor Homologue-1, LRH-1, Bound to DLPC and a Fragment of TIF-2 | | Descriptor: | DIUNDECYL PHOSPHATIDYL CHOLINE, Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2, ... | | Authors: | Musille, P.M, Ortlund, E.A. | | Deposit date: | 2012-02-10 | | Release date: | 2012-04-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Antidiabetic phospholipid-nuclear receptor complex reveals the mechanism for phospholipid-driven gene regulation.
Nat.Struct.Mol.Biol., 19, 2012
|
|
5W7Q
 
 | |
5W75
 
 | |
5W76
 
 | | Crystal Structure of Reconstructed Bacterial Elongation Factor Node 168 | | Descriptor: | Ancestral Elogation Factor N153, DI(HYDROXYETHYL)ETHER, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ortlund, E.A. | | Deposit date: | 2017-06-19 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Structural and Dynamics Comparison of Thermostability in Ancient, Modern, and Consensus Elongation Factor Tus.
Structure, 26, 2018
|
|
