6K5F
 
 | | Crystal structure of the CLC-ec1 deltaNC in presence of 200 mM NaBr | | Descriptor: | BROMIDE ION, Fab fragment, heavy chain, ... | | Authors: | Park, K, Lim, H.H. | | Deposit date: | 2019-05-28 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.203 Å) | | Cite: | Mutation of external glutamate residue reveals a new intermediate transport state and anion binding site in a CLC Cl-/H+antiporter.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6K5I
 
 | |
6K5A
 
 | |
6K5D
 
 | |
7CVT
 
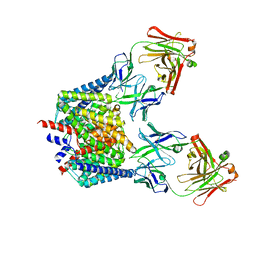 | | Crystal structure of the C85A/L194A/H234C mutant CLC-ec1 with Fab fragment | | Descriptor: | CHLORIDE ION, H(+)/Cl(-) exchange transporter ClcA, antibody Fab fragment heavy chain, ... | | Authors: | Park, K, Mersch, K, Robertson, J, Lim, H.-H. | | Deposit date: | 2020-08-27 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Altering CLC stoichiometry by reducing non-polar side-chains at the dimerization interface.
J.Mol.Biol., 433, 2021
|
|
7CVS
 
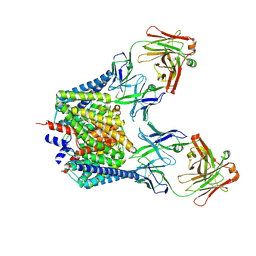 | | Crystal structure of the C85A/L194A mutant CLC-ec1 with Fab fragment | | Descriptor: | CHLORIDE ION, H(+)/Cl(-) exchange transporter ClcA, antibody Fab fragment heavy chain, ... | | Authors: | Park, K, Mersch, K, Robertson, J, Lim, H.-H. | | Deposit date: | 2020-08-27 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Altering CLC stoichiometry by reducing non-polar side-chains at the dimerization interface.
J.Mol.Biol., 433, 2021
|
|
3G42
 
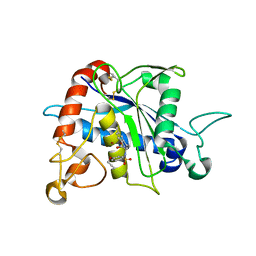 | | Crystal Structure of TACE with Tryptophan Sulfonamide Derivative Inhibitor | | Descriptor: | ADAM 17, N-{[4-(but-2-yn-1-yloxy)phenyl]sulfonyl}-5-methyl-D-tryptophan, ZINC ION | | Authors: | Xu, W, Park, K, Gopalsamy, A, Aplasca, A, Zhang, Y.H, Levin, J.I. | | Deposit date: | 2009-02-03 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synthesis and activity of tryptophan sulfonamide derivatives as novel non-hydroxamate TNF-alpha converting enzyme (TACE) inhibitors.
Bioorg.Med.Chem., 17, 2009
|
|
6AD8
 
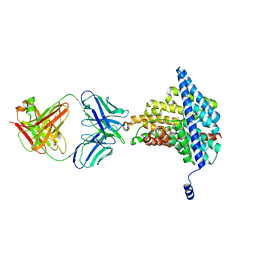 | | Crystal structure of the E148D mutant CLC-ec1 in 50 mM bromide | | Descriptor: | BROMIDE ION, H(+)/Cl(-) exchange transporter ClcA, antibody Fab fragment heavy chain, ... | | Authors: | Lim, H.-H, Park, K. | | Deposit date: | 2018-07-31 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Mutation of external glutamate residue reveals a new intermediate transport state and anion binding site in a CLC Cl-/H+antiporter.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6ADB
 
 | | Crystal structure of the E148N mutant CLC-ec1 in 20mM bromide | | Descriptor: | BROMIDE ION, H(+)/Cl(-) exchange transporter ClcA, antibody Fab fragment, ... | | Authors: | Lim, H.-H, Park, K. | | Deposit date: | 2018-07-31 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.692 Å) | | Cite: | Mutation of external glutamate residue reveals a new intermediate transport state and anion binding site in a CLC Cl-/H+antiporter.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6AD7
 
 | | Crystal structure of the E148D mutant CLC-ec1 in 20 mM bromide | | Descriptor: | BROMIDE ION, H(+)/Cl(-) exchange transporter ClcA, antibody Fab fragment heavy chain, ... | | Authors: | Lim, H.-H, Park, K. | | Deposit date: | 2018-07-31 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Mutation of external glutamate residue reveals a new intermediate transport state and anion binding site in a CLC Cl-/H+antiporter.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6ADC
 
 | |
6ADA
 
 | | Crystal structure of the E148D mutant CLC-ec1 in 200mM bromide | | Descriptor: | BROMIDE ION, H(+)/Cl(-) exchange transporter ClcA, antibody Fab fragment, ... | | Authors: | Lim, H.-H, Park, K. | | Deposit date: | 2018-07-31 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.153 Å) | | Cite: | Mutation of external glutamate residue reveals a new intermediate transport state and anion binding site in a CLC Cl-/H+antiporter.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7OCB
 
 | | Crystal structure of Spindlin1 in complex with the inhibitor XY49-92B | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 7-[3-(1,3-dihydroisoindol-2-yl)propoxy]-2N-[2-(dimethylamino)ethyl]-6-methoxy-4N-(1-propan-2-ylpiperidin-4-yl)quinazoline-2,4-diamine, CHLORIDE ION, ... | | Authors: | Johansson, C, Krojer, T, Park, K, Xiong, Y, Jin, J, Oppermann, U. | | Deposit date: | 2021-04-26 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal structure of Spindlin1 in complex with the inhibitor XY49-92B
To Be Published
|
|
8YBE
 
 | | Cryo-EM structure of Maltose Binding Protein | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yoo, Y, Park, K, Kim, H. | | Deposit date: | 2024-02-13 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Atomic resolution structure of MBP using Cryo-EM
To Be Published
|
|
4PSJ
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium (NESG) Target OR464. | | Descriptor: | OR464 | | Authors: | Vorobiev, S, Parmeggiani, F, Seetharaman, J, Huang, P.-S, Janjua, H, Xiao, R, Maglaqui, M, Park, K, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-03-07 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Crystal Structure of Engineered Protein OR464.
To be Published
|
|
4PQ8
 
 | | Crystal Structure of Engineered Protein, Northeast Structural Genomics Consortium Target OR465 | | Descriptor: | CHLORIDE ION, DESIGNED PROTEIN OR465 | | Authors: | Vorobiev, S, Parmeggiani, F, Seetharaman, J, Janjua, H, Xiao, R, Maglaqui, M, Park, K, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-02-28 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.833 Å) | | Cite: | Crystal Structure of Engineered Protein OR465.
To be Published
|
|
5ZWV
 
 | | Structural Basis for the Enantioselectivity of Est-Y29 toward (S)-ketoprofen | | Descriptor: | Est-Y29 | | Authors: | Ngo, D.T, Oh, C, Park, K, Nguyen, L, Byun, H.M, Kim, S, Yoon, S, Ryu, Y, Ryu, B.H, Kim, T.D, Yang, J.W. | | Deposit date: | 2018-05-17 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural Basis for the Enantioselectivity of Esterase Est-Y29 toward (S)-Ketoprofen
Acs Catalysis, 9, 2019
|
|
5ZWR
 
 | | Structural Basis for the Enantioselectivity of Est-Y29 toward (S)-ketoprofen | | Descriptor: | (2S)-2-[3-(benzenecarbonyl)phenyl]propanoic acid, Est-Y29, GLYCEROL | | Authors: | Ngo, D.T, Oh, C, Park, K, Nguyen, L, Byun, H.M, Kim, S, Yoon, S, Ryu, Y, Ryu, B.H, Kim, T.D, Kim, K.K. | | Deposit date: | 2018-05-16 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural Basis for the Enantioselectivity of Esterase Est-Y29 toward (S)-Ketoprofen
Acs Catalysis, 9, 2019
|
|
5ZWQ
 
 | | Structural Basis for the Enantioselectivity of Est-Y29 toward (S)-ketoprofen | | Descriptor: | Est-Y29, GLYCEROL, ethyl (2S)-2-[3-(benzenecarbonyl)phenyl]propanoate | | Authors: | Ngo, D.T, Oh, C, Park, K, Nguyen, L, Byun, H.M, Kim, S, Yoon, S, Ryu, Y, Ryu, B.H, Kim, T.D, Yang, J.W, Kim, K.K. | | Deposit date: | 2018-05-16 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Structural Basis for the Enantioselectivity of Esterase Est-Y29 toward (S)-Ketoprofen
Acs Catalysis, 9, 2019
|
|
4R58
 
 | |
4R5C
 
 | |
4R6F
 
 | |
5GV5
 
 | | Crystal structure of Candida antarctica Lipase B with active Ser105 modified with a phosphonate inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase B, ... | | Authors: | Park, S.Y, Lee, H. | | Deposit date: | 2016-09-02 | | Release date: | 2017-09-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structural and Experimental Evidence for the Enantiomeric Recognition toward a Bulky sec-Alcohol by Candida antarctica Lipase B
Acs Catalysis, 6, 2016
|
|
4BBW
 
 | | The crystal structure of Sialidase VPI 5482 (BTSA) from Bacteroides thetaiotaomicron | | Descriptor: | SIALIDASE (NEURAMINIDASE) | | Authors: | Park, K.-H, Song, H.-N, Jung, T.-Y, Lee, M.-H, Woo, E.-J. | | Deposit date: | 2012-09-28 | | Release date: | 2013-08-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Biochemical Characterization of the Broad Substrate Specificity of Bacteroides Thetaiotaomicron Commensal Sialidase.
Biochim.Biophys.Acta, 1834, 2013
|
|
4R6J
 
 | |
