5KSC
 
 | | E166A/R274N/R276N Toho-1 Beta-lactamase aztreonam acyl-enzyme intermediate | | Descriptor: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, Beta-lactamase Toho-1 | | Authors: | Vandavasi, V.G, Langan, P.S, Weiss, K, Parks, J.M, Cooper, J.B, Ginell, S.L, Coates, L. | | Deposit date: | 2016-07-08 | | Release date: | 2016-11-09 | | Last modified: | 2019-12-04 | | Method: | NEUTRON DIFFRACTION (2.1 Å) | | Cite: | Active-Site Protonation States in an Acyl-Enzyme Intermediate of a Class A beta-Lactamase with a Monobactam Substrate.
Antimicrob. Agents Chemother., 61, 2017
|
|
4XQD
 
 | | X-ray structure analysis of xylanase-WT at pH4.0 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-19 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XQW
 
 | | X-ray structure analysis of xylanase-N44E with MES at pH6.0 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-20 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XPV
 
 | | Neutron and X-ray structure analysis of xylanase: N44D at pH6 | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-18 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XQ4
 
 | | X-ray structure analysis of xylanase - N44D | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-19 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5G18
 
 | | Direct Observation of Active-site Protonation States in a Class A beta lactamase with a monobactam substrate | | Descriptor: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, BETA-LACTAMASE CTX-M-97, SULFATE ION | | Authors: | Vandavasi, V.G, Weiss, K.L, Parks, J.M, Cooper, J.B, Ginell, S.L, Coates, L. | | Deposit date: | 2016-03-23 | | Release date: | 2016-11-09 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Active-Site Protonation States in an Acyl-Enzyme Intermediate of a Class A beta-Lactamase with a Monobactam Substrate.
Antimicrob. Agents Chemother., 61, 2017
|
|
6C7A
 
 | | Conformational Changes in a Class A Beta lactamase that Prime it for Catalysis | | Descriptor: | Beta-lactamase Toho-1, SULFATE ION | | Authors: | Coates, L, Langan, P.S, Vandavasi, V.G, Cooper, S.J, Weiss, K.L, Ginell, S.L, Parks, J.M. | | Deposit date: | 2018-01-22 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Substrate Binding Induces Conformational Changes in a Class A Beta-lactamase That Prime It for Catalysis
Acs Catalysis, 8, 2018
|
|
6C78
 
 | | Substrate Binding Induces Conformational Changes In A Class A Beta Lactamase That Primes It For Catalysis | | Descriptor: | Beta-lactamase Toho-1 | | Authors: | Langan, P.S, Vandavasi, V.G, Cooper, S.J, Weiss, K.L, Ginell, S.L, Parks, J.M, Coates, L. | | Deposit date: | 2018-01-22 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-13 | | Method: | NEUTRON DIFFRACTION (1.75 Å) | | Cite: | Substrate Binding Induces Conformational Changes in a Class A Beta-lactamase That Prime It for Catalysis
Acs Catalysis, 8, 2018
|
|
6C79
 
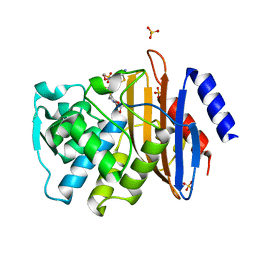 | | Conformational Changes in a Class A Beta lactamase that Prime it for Catalysis | | Descriptor: | (6R,7R)-3-(acetyloxymethyl)-7-[[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-methoxyimino-ethanoyl]amino]-8-oxo-5-thia-1-azabicy clo[4.2.0]oct-2-ene-2-carboxylic acid, Beta-lactamase Toho-1, SULFATE ION | | Authors: | Coates, L, Langan, P.S, Vandavasi, V.G, Cooper, S.J, Weiss, K.L, Ginell, S.L, Parks, J.M. | | Deposit date: | 2018-01-22 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Substrate Binding Induces Conformational Changes in a Class A Beta-lactamase That Prime It for Catalysis
Acs Catalysis, 8, 2018
|
|
8EZC
 
 | |
8EYP
 
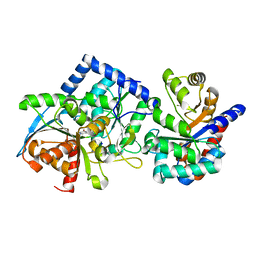 | | Joint X-ray/neutron structure of Salmonella typhimurium tryptophan synthase internal aldimine from microgravity-grown crystal | | Descriptor: | SODIUM ION, Tryptophan synthase alpha chain, Tryptophan synthase beta chain | | Authors: | Drago, V.N, Kovalevsky, A, Blakeley, M.P, Forsyth, V.T, Mueser, T.C. | | Deposit date: | 2022-10-28 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-28 | | Method: | NEUTRON DIFFRACTION (1.8 Å), X-RAY DIFFRACTION | | Cite: | Neutron diffraction from a microgravity-grown crystal reveals the active site hydrogens of the internal aldimine form of tryptophan synthase
Cell Rep Phys Sci, 5, 2024
|
|
8EYS
 
 | |
5KWJ
 
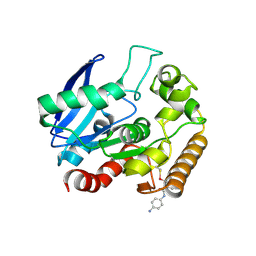 | | M.tb Ag85C modified at C209 by amino-ebselen | | Descriptor: | Diacylglycerol acyltransferase/mycolyltransferase Ag85C, ~{N}-(4-aminophenyl)-2-selanyl-benzamide | | Authors: | Goins, C.M, Ronning, D.R. | | Deposit date: | 2016-07-18 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Exploring Covalent Allosteric Inhibition of Antigen 85C from Mycobacterium tuberculosis by Ebselen Derivatives.
ACS Infect Dis, 3, 2017
|
|
5KWI
 
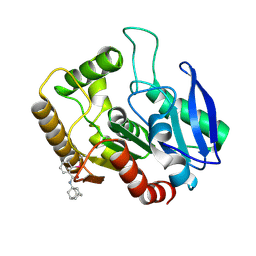 | | M.tb Ag85C modified at C209 by adamantyl-ebselen | | Descriptor: | Diacylglycerol acyltransferase/mycolyltransferase Ag85C, ~{N}-(1-adamantyl)-2-selanyl-benzamide | | Authors: | Goins, C.M, Ronning, D.R. | | Deposit date: | 2016-07-18 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Exploring Covalent Allosteric Inhibition of Antigen 85C from Mycobacterium tuberculosis by Ebselen Derivatives.
ACS Infect Dis, 3, 2017
|
|
7S3K
 
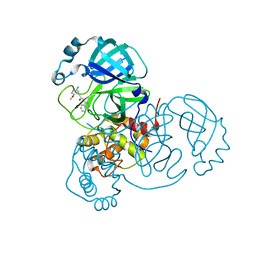 | |
7S4B
 
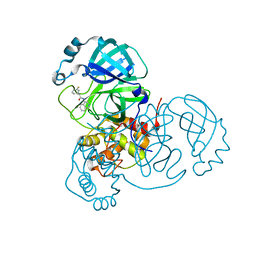 | |
7S3S
 
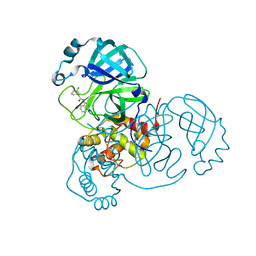 | |
2A2K
 
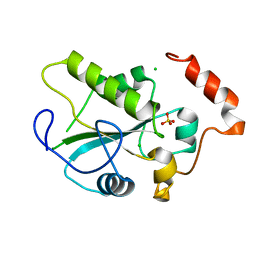 | | Crystal Structure of an active site mutant, C473S, of Cdc25B Phosphatase Catalytic Domain | | Descriptor: | CHLORIDE ION, M-phase inducer phosphatase 2, SULFATE ION | | Authors: | Sohn, J, Parks, J, Buhrman, G, Brown, P, Kristjansdottir, K, Safi, A, Yang, W, Edelsbrunner, H, Rudolph, J. | | Deposit date: | 2005-06-22 | | Release date: | 2006-01-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Experimental Validation of the Docking Orientation of Cdc25 with Its Cdk2-CycA Protein Substrate.
Biochemistry, 44, 2005
|
|
5E5J
 
 | | Joint X-ray/neutron structure of HIV-1 protease triple mutant (V32I,I47V,V82I) with darunavir at pH 6.0 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | Authors: | Kovalevsky, A.Y, Gerlits, O.O. | | Deposit date: | 2015-10-08 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | NEUTRON DIFFRACTION (1.85 Å), X-RAY DIFFRACTION | | Cite: | Long-Range Electrostatics-Induced Two-Proton Transfer Captured by Neutron Crystallography in an Enzyme Catalytic Site.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5E5K
 
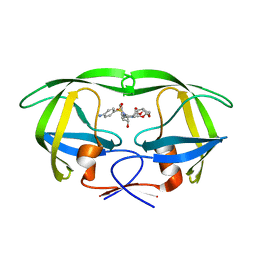 | | Joint X-ray/neutron structure of HIV-1 protease triple mutant (V32I,I47V,V82I) with darunavir at pH 4.3 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, HIV-1 protease | | Authors: | Kovalevsky, A.Y, Das, A. | | Deposit date: | 2015-10-08 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | NEUTRON DIFFRACTION (1.75 Å), X-RAY DIFFRACTION | | Cite: | Long-Range Electrostatics-Induced Two-Proton Transfer Captured by Neutron Crystallography in an Enzyme Catalytic Site.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
4QE5
 
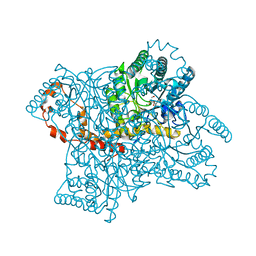 | |
4QE4
 
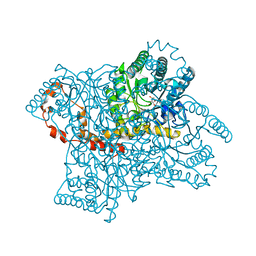 | |
4QEH
 
 | |
4QE1
 
 | |
4QDP
 
 | | Joint X-ray and neutron structure of Streptomyces rubiginosus D-xylose isomerase in complex with two Cd2+ ions and cyclic beta-L-arabinose | | Descriptor: | CADMIUM ION, Xylose isomerase, beta-L-arabinopyranose | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2014-05-14 | | Release date: | 2014-09-03 | | Last modified: | 2024-02-28 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | L-Arabinose Binding, Isomerization, and Epimerization by D-Xylose Isomerase: X-Ray/Neutron Crystallographic and Molecular Simulation Study.
Structure, 22, 2014
|
|
