3E64
 
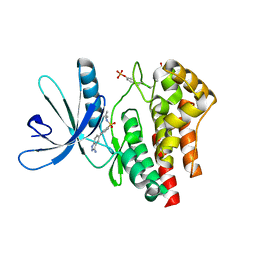 | | Fragment based discovery of JAK-2 inhibitors | | Descriptor: | 4-(3-amino-1H-indazol-5-yl)-N-tert-butylbenzenesulfonamide, Tyrosine-protein kinase JAK2 | | Authors: | Antonysamy, S, Fang, W, Hirst, G, Park, F, Russell, M, Smyth, L, Sprengeler, P, Stappenbeck, F, Steensma, R, Thompson, D.A, Wilson, M, Wong, M, Zhang, A, Zhang, F. | | Deposit date: | 2008-08-14 | | Release date: | 2008-10-14 | | Last modified: | 2012-02-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment-based discovery of JAK-2 inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3E63
 
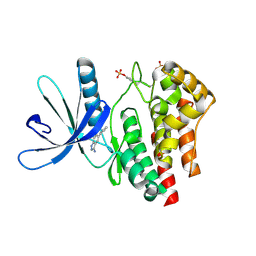 | | Fragment based discovery of JAK-2 inhibitors | | Descriptor: | 5-phenyl-1H-indazol-3-amine, Tyrosine-protein kinase JAK2 | | Authors: | Antonysamy, S, Fang, W, Hirst, G, Park, F, Russell, M, Smyth, L, Sprengeler, P, Stappenbeck, F, Steensma, R, Thompson, D.A, Wilson, M, Wong, M, Zhang, A, Zhang, F. | | Deposit date: | 2008-08-14 | | Release date: | 2008-10-14 | | Last modified: | 2012-02-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment-based discovery of JAK-2 inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3E62
 
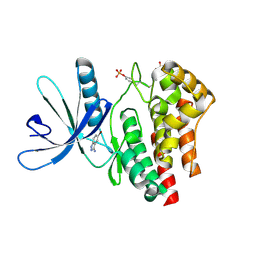 | | Fragment based discovery of JAK-2 inhibitors | | Descriptor: | 5-bromo-1H-indazol-3-amine, Tyrosine-protein kinase JAK2 | | Authors: | Antonysamy, S, Fang, W, Hirst, G, Park, F, Russell, M, Smyth, L, Sprengeler, P, Stappenbeck, F, Steensma, R, Thompson, D.A, Wilson, M, Wong, M, Zhang, A, Zhang, F. | | Deposit date: | 2008-08-14 | | Release date: | 2008-10-14 | | Last modified: | 2012-02-08 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | Fragment-based discovery of JAK-2 inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
5H7O
 
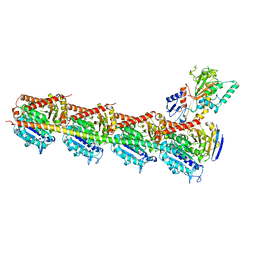 | | Crystal structure of DJ-101 in complex with tubulin protein | | Descriptor: | 2-(1H-indol-4-yl)-4-(3,4,5-trimethoxyphenyl)-1H-imidazo[4,5-c]pyridine, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Arnst, K, Wang, Y, Hwang, D.-J, Xue, Y, Costello, T, Hamilton, D, Chen, Q, Yang, J, Park, F, Dalton, J.T, Miller, D.D, Li, W. | | Deposit date: | 2016-11-20 | | Release date: | 2017-12-13 | | Last modified: | 2022-10-12 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Potent, Metabolically Stable Tubulin Inhibitor Targets the Colchicine Binding Site and Overcomes Taxane Resistance.
Cancer Res., 78, 2018
|
|
3RU0
 
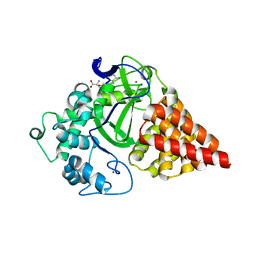 | | Cocrystal structure of human SMYD3 with inhibitor Sinefungin bound | | Descriptor: | SET and MYND domain-containing protein 3, SINEFUNGIN, ZINC ION | | Authors: | Foreman, K.W, Brown, M, Park, F, Emtage, S, Harriss, J, Das, C, Zhu, L, Crew, A, Arnold, L, Shaaban, S, Tucker, P. | | Deposit date: | 2011-05-04 | | Release date: | 2011-05-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Structural and Functional Profiling of the Human Histone Methyltransferase SMYD3.
Plos One, 6, 2011
|
|
1VHN
 
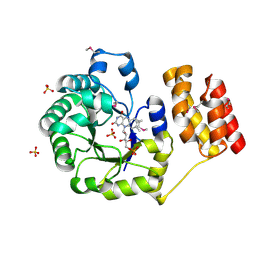 | |
1VI7
 
 | | Crystal structure of an hypothetical protein | | Descriptor: | Hypothetical protein yigZ | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of YIGZ, a conserved hypothetical protein from Escherichia coli k12 with a novel fold
Proteins, 55, 2004
|
|
1O61
 
 | | Crystal structure of a PLP-dependent enzyme with PLP | | Descriptor: | ACETATE ION, PYRIDOXAL-5'-PHOSPHATE, aminotransferase | | Authors: | Structural GenomiX | | Deposit date: | 2003-10-23 | | Release date: | 2003-11-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1O68
 
 | |
1O6D
 
 | | Crystal structure of a hypothetical protein | | Descriptor: | Hypothetical UPF0247 protein TM0844 | | Authors: | Structural GenomiX | | Deposit date: | 2003-11-03 | | Release date: | 2003-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1O69
 
 | | Crystal structure of a PLP-dependent enzyme | | Descriptor: | (2-AMINO-4-FORMYL-5-HYDROXY-6-METHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, BETA-MERCAPTOETHANOL, aminotransferase | | Authors: | Structural GenomiX | | Deposit date: | 2003-10-23 | | Release date: | 2003-11-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1O64
 
 | |
1O62
 
 | |
1O65
 
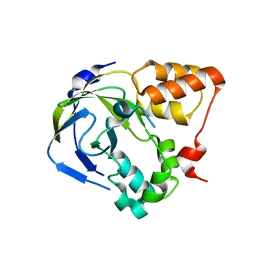 | | Crystal structure of an hypothetical protein | | Descriptor: | Hypothetical protein yiiM | | Authors: | Structural GenomiX | | Deposit date: | 2003-10-23 | | Release date: | 2003-11-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1LEI
 
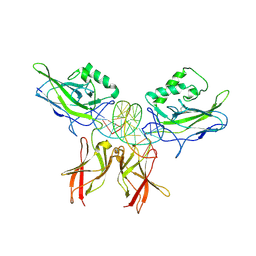 | | The kB DNA sequence from the HLV-LTR functions as an allosteric regulator of HIV transcription | | Descriptor: | 5'-D(*CP*TP*CP*AP*GP*GP*GP*AP*AP*AP*GP*TP*AP*CP*AP*GP*A)-3', 5'-D(*TP*CP*TP*GP*5ITP*AP*CP*5ITP*5ITP*5ITP*CP*CP*CP*TP*GP*AP*G)-3', NUCLEAR FACTOR NF-KAPPA-B P50 SUBUNIT, ... | | Authors: | Chen-Park, F, Huang, D.B, Ghosh, G. | | Deposit date: | 2002-04-09 | | Release date: | 2003-04-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The kB DNA sequence from the HIV Long Terminal Repeat functions as an allosteric regulator of HIV transcription
J.Biol.Chem., 277, 2002
|
|
1O63
 
 | |
1O6C
 
 | |
1O6B
 
 | |
1O66
 
 | |
1O60
 
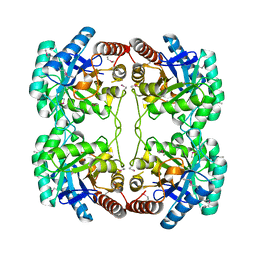 | | Crystal structure of KDO-8-phosphate synthase | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase | | Authors: | Structural GenomiX | | Deposit date: | 2003-10-23 | | Release date: | 2003-11-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1O67
 
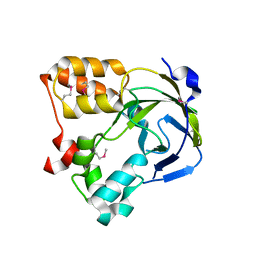 | | Crystal structure of an hypothetical protein | | Descriptor: | Hypothetical protein yiiM | | Authors: | Structural GenomiX | | Deposit date: | 2003-10-23 | | Release date: | 2003-11-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VGU
 
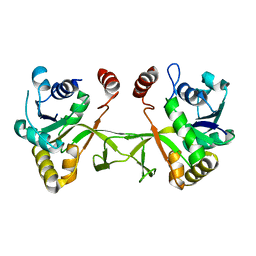 | |
1VH0
 
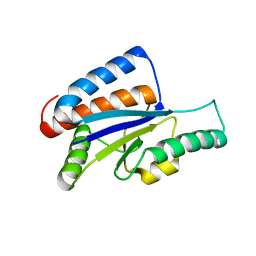 | | Crystal structure of a hypothetical protein | | Descriptor: | Hypothetical UPF0247 protein SAV0024/SA0023 | | Authors: | Structural GenomiX | | Deposit date: | 2003-11-03 | | Release date: | 2003-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VHL
 
 | |
1VI1
 
 | |
