7PVB
 
 | |
6ZZE
 
 | |
6ZZF
 
 | |
2MJ0
 
 | |
6ZSS
 
 | |
6IB6
 
 | | Solution structure of the water-soluble LU-domain of human Lypd6 protein | | Descriptor: | Ly6/PLAUR domain-containing protein 6 | | Authors: | Tsarev, A.V, Kulbatskii, D.S, Paramonov, A.S, Lyukmanova, E.N, Shenkarev, Z.O. | | Deposit date: | 2018-11-29 | | Release date: | 2019-12-18 | | Last modified: | 2021-01-13 | | Method: | SOLUTION NMR | | Cite: | Structural Diversity and Dynamics of Human Three-Finger Proteins Acting on Nicotinic Acetylcholine Receptors.
Int J Mol Sci, 21, 2020
|
|
6ZSO
 
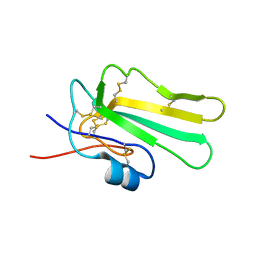 | | Solution structure of the water-soluble LU-domain of human Lypd6b protein | | Descriptor: | Ly6/PLAUR domain-containing protein 6B | | Authors: | Tsarev, A.V, Kulbatskii, D.S, Paramonov, A.S, Lyukmanova, E.N, Shenkarev, Z.O. | | Deposit date: | 2020-07-16 | | Release date: | 2021-01-13 | | Method: | SOLUTION NMR | | Cite: | Structural Diversity and Dynamics of Human Three-Finger Proteins Acting on Nicotinic Acetylcholine Receptors.
Int J Mol Sci, 21, 2020
|
|
2N99
 
 | | Solution structure of the SLURP-2, a secreted isoform of Lynx1 | | Descriptor: | Ly-6/neurotoxin-like protein 1 | | Authors: | Paramonov, A.S, Shenkarev, Z.O, Lyukmanova, E.N, Arseniev, A.S. | | Deposit date: | 2015-11-11 | | Release date: | 2016-09-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Secreted Isoform of Human Lynx1 (SLURP-2): Spatial Structure and Pharmacology of Interactions with Different Types of Acetylcholine Receptors.
Sci Rep, 6, 2016
|
|
6HN9
 
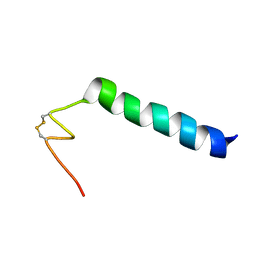 | | Nicomicin-1 -- Novel antimicrobial peptides from the Arctic polychaeta Nicomache minor provide new molecular insight into biological role of the BRICHOS domain | | Descriptor: | Nicomicin-1 | | Authors: | Panteleev, P.V, Tsarev, A.V, Bolosov, I.A, Paramonov, A.S, Marggraf, M.B, Sychev, S.V, Shenkarev, Z.O, Ovchinnikova, T.V. | | Deposit date: | 2018-09-14 | | Release date: | 2018-11-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Novel Antimicrobial Peptides from the Arctic PolychaetaNicomache minorProvide New Molecular Insight into Biological Role of the BRICHOS Domain.
Mar Drugs, 16, 2018
|
|
8B4R
 
 | | Antimicrobial peptide capitellacin from polychaeta Capitella teleta in DPC (dodecylphosphocholine) micelles, monomeric form | | Descriptor: | BRICHOS domain-containing protein | | Authors: | Mironov, P.A, Reznikova, O.V, Paramonov, A.S, Shenkarev, Z.O. | | Deposit date: | 2022-09-21 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Dimerization of the beta-Hairpin Membrane-Active Cationic Antimicrobial Peptide Capitellacin from Marine Polychaeta: An NMR Structural and Thermodynamic Study.
Biomolecules, 14, 2024
|
|
8C5J
 
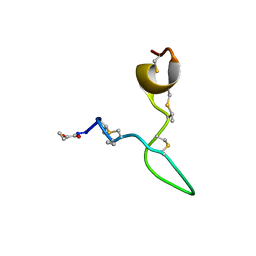 | | Spatial structure of Lch-alpha peptide from two-component lantibiotic system Lichenicidin VK21 | | Descriptor: | Lantibiotic lichenicidin VK21 A1 | | Authors: | Mineev, K.S, Paramonov, A.S, Arseniev, A.S, Ovchinnikova, T.V, Shenkarev, Z.O. | | Deposit date: | 2023-01-09 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Specific Binding of the alpha-Component of the Lantibiotic Lichenicidin to the Peptidoglycan Precursor Lipid II Predetermines Its Antimicrobial Activity.
Int J Mol Sci, 24, 2023
|
|
2L8X
 
 | |
2M1F
 
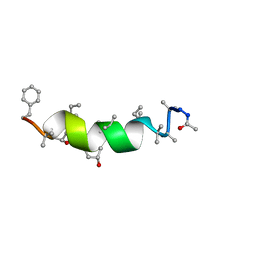 | | NMR Structure of Antiamoebin I (peptaibol antibiotic) bound to DMPC/DHPC bicelles | | Descriptor: | Antiamoebin I | | Authors: | Shenkarev, Z.O, Paramonov, A.S, Gizatullina, A.K. | | Deposit date: | 2012-11-27 | | Release date: | 2012-12-12 | | Last modified: | 2013-10-02 | | Method: | SOLUTION NMR | | Cite: | Peptaibol antiamoebin I: spatial structure, backbone dynamics, interaction with bicelles and lipid-protein nanodiscs, and pore formation in context of barrel-stave model.
Chem.Biodivers., 10, 2013
|
|
2MQU
 
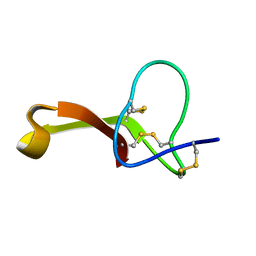 | | Spatial structure of Hm-3, a membrane-active spider toxin affecting sodium channels | | Descriptor: | Neurotoxin Hm-3 | | Authors: | Myshkin, M.Y, Shenkarev, Z.O, Paramonov, A.S, Berkut, A.A, Grishin, E.V, Vassilevski, A.A. | | Deposit date: | 2014-06-27 | | Release date: | 2014-11-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of membrane-active toxin from crab spider Heriaeus melloteei suggests parallel evolution of sodium channel gating modifiers in Araneomorphae and Mygalomorphae.
J. Biol. Chem., 290, 2015
|
|
8B4S
 
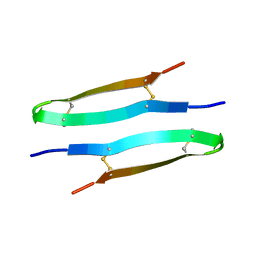 | | Antimicrobial peptide capitellacin from polychaeta Capitella teleta in DPC (dodecylphosphocholine) micelles, dimeric form | | Descriptor: | BRICHOS domain-containing protein | | Authors: | Mironov, P.A, Reznikova, O.V, Paramonov, A.S, Shenkarev, Z.O. | | Deposit date: | 2022-09-21 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Dimerization of the beta-Hairpin Membrane-Active Cationic Antimicrobial Peptide Capitellacin from Marine Polychaeta: An NMR Structural and Thermodynamic Study.
Biomolecules, 14, 2024
|
|
8BWB
 
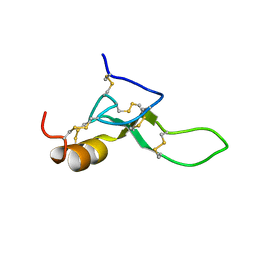 | | Spider toxin Pha1b (PnTx3-6) from Phoneutria nigriventer targeting CaV2.x calcium channels and TRPA1 channel | | Descriptor: | Omega-ctenitoxin-Pn4a | | Authors: | Mironov, P.A, Chernaya, E.M, Paramonov, A.S, Shenkarev, Z.O. | | Deposit date: | 2022-12-06 | | Release date: | 2023-06-21 | | Last modified: | 2023-07-05 | | Method: | SOLUTION NMR | | Cite: | Recombinant Production, NMR Solution Structure, and Membrane Interaction of the Ph alpha 1 beta Toxin, a TRPA1 Modulator from the Brazilian Armed Spider Phoneutria nigriventer .
Toxins, 15, 2023
|
|
2N81
 
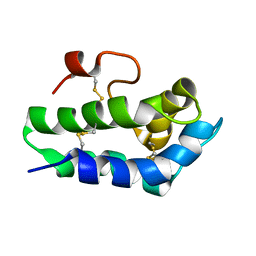 | | Solution Structure of Lipid Transfer Protein From Pea Pisum Sativum | | Descriptor: | Lipid Transfer Protein | | Authors: | Paramonov, A.S, Rumynskiy, E.I, Bogdanov, I.V, Finkina, E.I, Melnikova, D.N, Ovchinnikova, T.V, Shenkarev, Z.O, Arseniev, A.S. | | Deposit date: | 2015-09-30 | | Release date: | 2016-05-11 | | Last modified: | 2017-12-20 | | Method: | SOLUTION NMR | | Cite: | A novel lipid transfer protein from the pea Pisum sativum: isolation, recombinant expression, solution structure, antifungal activity, lipid binding, and allergenic properties.
BMC Plant Biol, 16
|
|
