5HI6
 
 | | The high resolution structure of dihydrofolate reductase from Yersinia pestis complex with methotrexate as closed form | | Descriptor: | CALCIUM ION, CHLORIDE ION, Dihydrofolate reductase, ... | | Authors: | Chang, C, Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-11 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | The high resolution structure of dihydrofolate reductase from Yersinia pestis complex with methotrexate as closed form
To Be Published
|
|
5CXD
 
 | | 1.75 Angstrom resolution crystal structure of the apo-form acyl-carrier-protein synthase (AcpS) (acpS; purification tag off) from Staphylococcus aureus subsp. aureus COL in the I4 space group | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Holo-[acyl-carrier-protein] synthase, ... | | Authors: | Halavaty, A.S, Minasov, G, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-07-28 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | 1.75 Angstrom resolution crystal structure of the apo-form acyl-carrier-protein synthase (AcpS) (acpS; purification tag off) from Staphylococcus aureus subsp. aureus COL in the I4 space group
To Be Published
|
|
3DZC
 
 | | 2.35 Angstrom resolution structure of WecB (VC0917), a UDP-N-acetylglucosamine 2-epimerase from Vibrio cholerae. | | Descriptor: | CALCIUM ION, CHLORIDE ION, UDP-N-acetylglucosamine 2-epimerase | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Kwon, K, Hasseman, J, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-07-29 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 2.35 Angstrom resolution structure of WecB (VC0917), a UDP-N-acetylglucosamine 2-epimerase from Vibrio cholerae.
TO BE PUBLISHED
|
|
5FDA
 
 | | The high resolution structure of apo form dihydrofolate reductase from Yersinia pestis at 1.55 A | | Descriptor: | CHLORIDE ION, Dihydrofolate reductase | | Authors: | Chang, C, Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-15 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | structure of dihydrofolate reductase from Yersinia pestis complex with
To Be Published
|
|
5HG0
 
 | | Crystal Structure of Pantoate-beta-alanine Ligase from Francisella tularensis complex with SAM | | Descriptor: | Pantothenate synthetase, S-ADENOSYLMETHIONINE | | Authors: | Chang, C, Maltseva, N, Kim, Y, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-07 | | Release date: | 2016-01-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Pantoate-beta-alanine Ligase from Francisella tularensis complex with SAM
To Be Published
|
|
3ECT
 
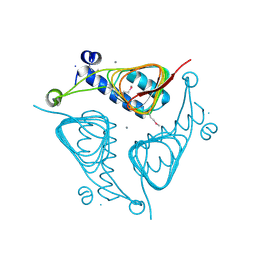 | | Crystal Structure of the Hexapeptide-Repeat Containing-Acetyltransferase VCA0836 from Vibrio cholerae | | Descriptor: | CALCIUM ION, Hexapeptide-repeat containing-acetyltransferase | | Authors: | Kim, Y, Maltseva, N, Kwon, K, Papazisi, L, Hasseman, J, Peterson, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-09-02 | | Release date: | 2008-09-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal Structure of the Hexapeptide-Repeat Containing-Acetyltransferase VCA0836 from Vibrio cholerae
To be Published
|
|
4FCU
 
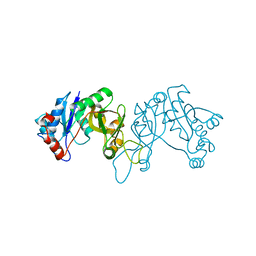 | | 1.9 Angstrom Crystal Structure of 3-deoxy-manno-octulosonate Cytidylyltransferase (kdsB) from Acinetobacter baumannii without His-Tag Bound to the Active Site | | Descriptor: | 3-deoxy-manno-octulosonate cytidylyltransferase | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-25 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom Crystal Structure of 3-deoxy-manno-octulosonate Cytidylyltransferase (kdsB) from Acinetobacter baumannii without His-Tag Bound to the Active Site.
TO BE PUBLISHED
|
|
3GEU
 
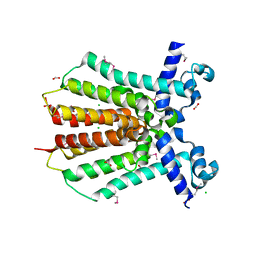 | | Crystal Structure of IcaR from Staphylococcus aureus, a member of the tetracycline repressor protein family | | Descriptor: | CHLORIDE ION, FORMIC ACID, Intercellular adhesion protein R, ... | | Authors: | Anderson, S.M, Brunzelle, J.S, Wawrzak, Z, Skarina, T, Papazisi, L, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-02-26 | | Release date: | 2009-03-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of IcaR from Staphylococcus aureus, a member of the tetracycline repressor protein family
To be Published
|
|
4GIB
 
 | | 2.27 Angstrom Crystal Structure of beta-Phosphoglucomutase (pgmB) from Clostridium difficile | | Descriptor: | Beta-phosphoglucomutase, GLYCINE, PHOSPHATE ION, ... | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Grimshaw, S, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-08 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | 2.27 Angstrom Crystal Structure of beta-Phosphoglucomutase (pgmB) from Clostridium difficile.
TO BE PUBLISHED
|
|
4GFQ
 
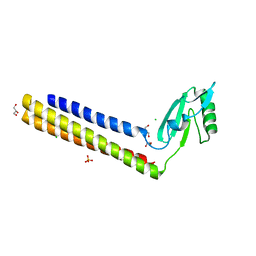 | | 2.65 Angstrom Resolution Crystal Structure of Ribosome Recycling Factor (frr) from Bacillus anthracis | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-03 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | 2.65 Angstrom Resolution Crystal Structure of Ribosome Recycling Factor (frr) from Bacillus anthracis
TO BE PUBLISHED
|
|
4GFP
 
 | | 2.7 Angstrom resolution structure of 3-phosphoshikimate 1-carboxyvinyltransferase (AroA) from Coxiella burnetii in a second conformational state | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, BETA-MERCAPTOETHANOL | | Authors: | Light, S.H, Minasov, G, Krishna, S.N, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-03 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 2.7 Angstrom resolution structure of 3-phosphoshikimate 1-carboxyvinyltransferase (AroA) from Coxiella burnetii in second conformational state
TO BE PUBLISHED
|
|
4GHJ
 
 | | 1.75 Angstrom Crystal Structure of Transcriptional Regulator ftom Vibrio vulnificus. | | Descriptor: | Probable transcriptional regulator | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Grimshaw, S, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-07 | | Release date: | 2012-08-15 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | 1.75 Angstrom Crystal Structure of Transcriptional Regulator ftom Vibrio vulnificus.
TO BE PUBLISHED
|
|
4H4N
 
 | | 1.1 Angstrom Crystal Structure of Hypothetical Protein BA_2335 from Bacillus anthracis | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Dubrovska, I, Winsor, J, Grimshaw, S, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-09-17 | | Release date: | 2012-09-26 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | 1.1 Angstrom Crystal Structure of Hypothetical Protein BA_2335 from Bacillus anthracis.
TO BE PUBLISHED
|
|
4H3D
 
 | | 1.95 Angstrom Crystal Structure of of Type I 3-Dehydroquinate Dehydratase (aroD) from Clostridium difficile with Covalent Modified Comenic Acid. | | Descriptor: | 3-dehydroquinate dehydratase, 5-hydroxy-6-methyl-4-oxo-4H-pyran-2-carboxylic acid, ACETATE ION, ... | | Authors: | Minasov, G, Light, S.H, Shuvalova, L, Duban, M.-E, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-09-13 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Crystal Structure of of Type I 3-Dehydroquinate Dehydratase (aroD) from Clostridium difficile with Covalent Modified Comenic Acid.
TO BE PUBLISHED
|
|
4JM7
 
 | | 1.82 Angstrom resolution crystal structure of holo-(acyl-carrier-protein) synthase (acpS) from Staphylococcus aureus | | Descriptor: | Holo-[acyl-carrier-protein] synthase | | Authors: | Halavaty, A.S, Minasov, G, Shuvalova, L, Dubrovska, I, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-13 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.824 Å) | | Cite: | Structural characterization and comparison of three acyl-carrier-protein synthases from pathogenic bacteria.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4DB3
 
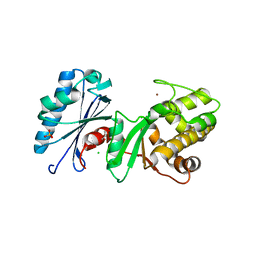 | | 1.95 Angstrom Resolution Crystal Structure of N-acetyl-D-glucosamine kinase from Vibrio vulnificus. | | Descriptor: | CHLORIDE ION, GLYCEROL, N-acetyl-D-glucosamine kinase, ... | | Authors: | Minasov, G, Wawrzak, Z, Onopriyenko, O, Skarina, T, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-01-13 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Resolution Crystal Structure of N-acetyl-D-glucosamine kinase from Vibrio vulnificus.
TO BE PUBLISHED
|
|
4DIB
 
 | | The crystal structure of glyceraldehyde-3-phosphate dehydrogenase from Bacillus anthracis str. Sterne | | Descriptor: | Glyceraldehyde 3-phosphate dehydrogenase, SULFATE ION | | Authors: | Nocek, B, Makowska-Grzyska, M, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-01-30 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The crystal structure of glyceraldehyde-3-phosphate dehydrogenase from Bacillus anthracis str. Sterne
To be Published
|
|
4E16
 
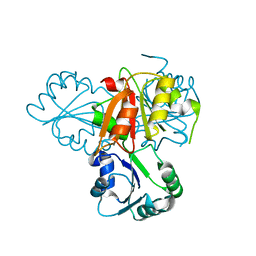 | | Precorrin-4 C(11)-methyltransferase from Clostridium difficile | | Descriptor: | precorrin-4 C(11)-methyltransferase | | Authors: | Osipiuk, J, Nocek, B, Makowska-Grzyska, M, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-05 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Precorrin-4 C(11)-methyltransferase from Clostridium difficile
To be Published
|
|
4EG2
 
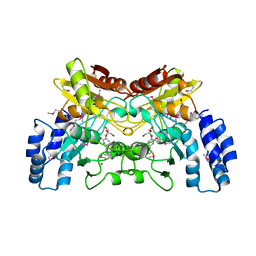 | | 2.2 Angstrom Crystal Structure of Cytidine deaminase from Vibrio cholerae in Complex with Zinc and Uridine | | Descriptor: | ACETATE ION, Cytidine deaminase, MAGNESIUM ION, ... | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Wang, Y, Grimshaw, S, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-30 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 2.2 Angstrom Crystal Structure of Cytidine deaminase from Vibrio cholerae in Complex with Zinc and Uridine.
TO BE PUBLISHED
|
|
3SD7
 
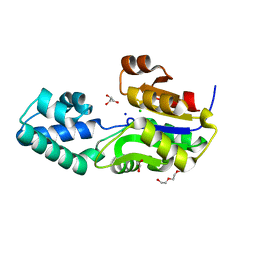 | | 1.7 Angstrom Resolution Crystal Structure of Putative Phosphatase from Clostridium difficile | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative phosphatase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-06-08 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7 Angstrom Resolution Crystal Structure of Putative Phosphatase from Clostridium difficile.
TO BE PUBLISHED
|
|
3SEF
 
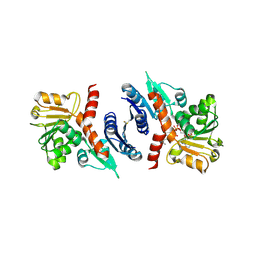 | | 2.4 Angstrom resolution crystal structure of shikimate 5-dehydrogenase (aroE) from Vibrio cholerae O1 biovar eltor str. N16961 in complex with shikimate and NADPH | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Shikimate 5-dehydrogenase | | Authors: | Halavaty, A.S, Light, S.H, Minasov, G, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-06-10 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4 Angstrom resolution crystal structure of shikimate 5-dehydrogenase (aroE) from Vibrio cholerae O1 biovar eltor str. N16961 in complex with shikimate and NADPH
To be Published
|
|
3SLH
 
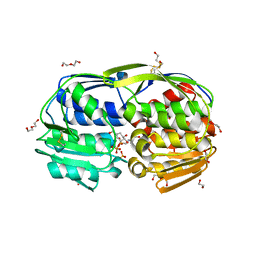 | | 1.70 Angstrom resolution structure of 3-phosphoshikimate 1-carboxyvinyltransferase (AroA) from Coxiella burnetii in complex with shikimate-3-phosphate and glyphosate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 1,2-ETHANEDIOL, 3-phosphoshikimate 1-carboxyvinyltransferase, ... | | Authors: | Light, S.H, Minasov, G, Filippova, E.V, Krishna, S.N, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-06-24 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.70 Angstrom resolution structure of 3-phosphoshikimate 1-carboxyvinyltransferase(AroA) from Coxiella burnetii in complex with shikimate-3-phosphate and glyphosate
To be Published
|
|
3T4E
 
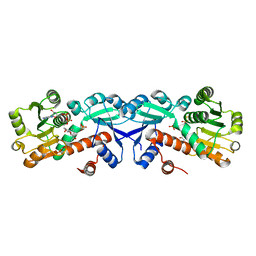 | | 1.95 Angstrom Crystal Structure of Shikimate 5-dehydrogenase (AroE) from Salmonella enterica subsp. enterica serovar Typhimurium in Complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION, Quinate/shikimate dehydrogenase | | Authors: | Minasov, G, Light, S.H, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-25 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Crystal Structure of Shikimate 5-dehydrogenase (AroE) from Salmonella enterica subsp. enterica serovar Typhimurium in Complex with NAD.
TO BE PUBLISHED
|
|
3TI2
 
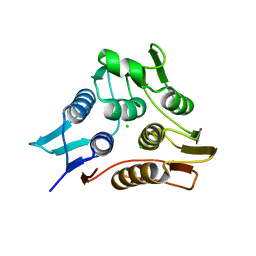 | | 1.90 Angstrom resolution crystal structure of N-terminal domain 3-phosphoshikimate 1-carboxyvinyltransferase from Vibrio cholerae | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, CHLORIDE ION, TETRAETHYLENE GLYCOL | | Authors: | Light, S.H, Minasov, G, Halavaty, A.S, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-08-19 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.90 Angstrom resolution crystal structure of N-terminal domain 3-phosphoshikimate 1-carboxyvinyltransferase from Vibrio cholerae
TO BE PUBLISHED
|
|
3TNL
 
 | | 1.45 Angstrom Crystal Structure of Shikimate 5-dehydrogenase from Listeria monocytogenes in Complex with Shikimate and NAD. | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Minasov, G, Light, S.H, Halavaty, A, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-01 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | 1.45 Angstrom Crystal Structure of Shikimate 5-dehydrogenase from Listeria monocytogenes in Complex with Shikimate and NAD.
TO BE PUBLISHED
|
|
