2R0I
 
 | |
1Y8G
 
 | |
5MVV
 
 | | Crystal structure of Plasmodium falciparum actin I- gelsolin segment 1 -CdATP complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-1, CADMIUM ION, ... | | Authors: | Panneerselvam, S, Kumpula, E.-P, Kursula, I, Burkhardt, A, Meents, A. | | Deposit date: | 2017-01-17 | | Release date: | 2017-07-12 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Rapid cadmium SAD phasing at the standard wavelength (1 angstrom ).
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5MYY
 
 | | Hen Egg-White Lysozyme (HEWL) cocrystallized in the presence of Cadmium sulphate | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Panneerselvam, S, Burkhardt, A, Meents, A. | | Deposit date: | 2017-01-30 | | Release date: | 2017-07-12 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Rapid cadmium SAD phasing at the standard wavelength (1 angstrom ).
Acta Crystallogr D Struct Biol, 73, 2017
|
|
1ZMW
 
 | |
1ZMU
 
 | |
1ZMV
 
 | |
2WZJ
 
 | | Catalytic and UBA domain of kinase MARK2/(Par-1) K82R, T208E double mutant | | Descriptor: | SERINE/THREONINE-PROTEIN KINASE MARK2 | | Authors: | Panneerselvam, S, Marx, A, Mandelkow, E.-M, Mandelkow, E. | | Deposit date: | 2009-11-30 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.786 Å) | | Cite: | Structure and Function of Polarity-Inducing Kinase Family Mark/Par-1 within the Branch of Ampk/Snf1-Related Kinases.
Faseb J., 24, 2010
|
|
4PL9
 
 | |
4X3B
 
 | | A micro-patterned silicon chip as sample holder for macromolecular crystallography experiments with minimal background scattering | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Roedig, P, Vartiainen, I, Duman, R, Panneerselvam, S, Stuebe, N, Lorbeer, O, Warmer, M, Sutton, G, Stuart, D.I, Weckert, E, David, C, Wagner, A, Meents, A. | | Deposit date: | 2014-11-28 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A micro-patterned silicon chip as sample holder for macromolecular crystallography experiments with minimal background scattering.
Sci Rep, 5, 2015
|
|
3FE3
 
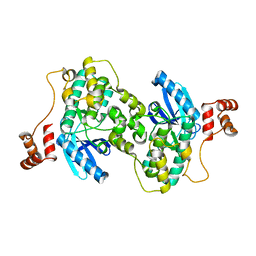 | |
2HAK
 
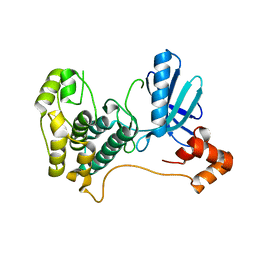 | | Catalytic and ubiqutin-associated domains of MARK1/PAR-1 | | Descriptor: | Serine/threonine-protein kinase MARK1 | | Authors: | Marx, A, Nugoor, C, Mueller, J, Panneerselvam, S, Mandelkow, E.-M, Mandelkow, E. | | Deposit date: | 2006-06-13 | | Release date: | 2006-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural variations in the catalytic and ubiquitin-associated domains of microtubule-associated protein/microtubule affinity regulating kinase (MARK) 1 and MARK2
J.Biol.Chem., 281, 2006
|
|
4X35
 
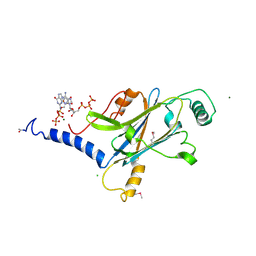 | | A micro-patterned silicon chip as sample holder for macromolecular crystallography experiments with minimal background scattering | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Roedig, P, Vartiainen, I, Duman, R, Panneerselvam, S, Stuebe, N, Lorbeer, O, Warmer, M, Sutton, G, Stuart, D.I, Weckert, E, David, C, Wagner, A, Meents, A. | | Deposit date: | 2014-11-27 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A micro-patterned silicon chip as sample holder for macromolecular crystallography experiments with minimal background scattering.
Sci Rep, 5, 2015
|
|
3P86
 
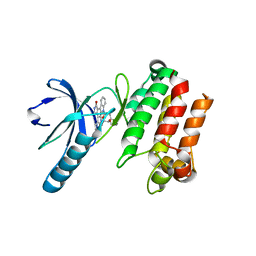 | |
3PPZ
 
 | |
4HGH
 
 | | Crystal structure of P450 BM3 5F5 heme domain variant complexed with styrene (dataset I) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bifunctional P-450/NADPH-P450 reductase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shehzad, A, Panneerselvam, S, Bocola, M, Mueller-Dieckmann, J, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2012-10-08 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | P450 BM3 crystal structures reveal the role of the charged surface residue Lys/Arg184 in inversion of enantioselective styrene epoxidation.
Chem.Commun.(Camb.), 49, 2013
|
|
4HGF
 
 | | Crystal structure of P450 BM3 5F5K heme domain variant complexed with styrene | | Descriptor: | Bifunctional P-450/NADPH-P450 reductase, CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Shehzad, A, Panneerselvam, S, Bocola, M, Mueller-Dieckmann, J, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2012-10-08 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | P450 BM3 crystal structures reveal the role of the charged surface residue Lys/Arg184 in inversion of enantioselective styrene epoxidation.
Chem.Commun.(Camb.), 49, 2013
|
|
4HGG
 
 | | Crystal structure of P450 BM3 5F5R heme domain variant complexed with styrene | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bifunctional P-450/NADPH-P450 reductase, GLYCEROL, ... | | Authors: | Shehzad, A, Panneerselvam, S, Bocola, M, Mueller-Dieckmann, J, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2012-10-08 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | P450 BM3 crystal structures reveal the role of the charged surface residue Lys/Arg184 in inversion of enantioselective styrene epoxidation.
Chem.Commun.(Camb.), 49, 2013
|
|
4HGJ
 
 | | Crystal structure of P450 BM3 5F5 heme domain variant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bifunctional P-450/NADPH-P450 reductase, GLYCEROL, ... | | Authors: | Shehzad, A, Panneerselvam, S, Bocola, M, Mueller-Dieckmann, J, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2012-10-08 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | P450 BM3 crystal structures reveal the role of the charged surface residue Lys/Arg184 in inversion of enantioselective styrene epoxidation.
Chem.Commun.(Camb.), 49, 2013
|
|
4HGI
 
 | | Crystal structure of P450 BM3 5F5 heme domain variant complexed with styrene (dataset II) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bifunctional P-450/NADPH-P450 reductase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shehzad, A, Panneerselvam, S, Bocola, M, Mueller-Dieckmann, J, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2012-10-08 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | P450 BM3 crystal structures reveal the role of the charged surface residue Lys/Arg184 in inversion of enantioselective styrene epoxidation.
Chem.Commun.(Camb.), 49, 2013
|
|
5E78
 
 | | Crystal structure of P450 BM3 heme domain variant complexed with Co(III)Sep | | Descriptor: | 1,3,6,8,10,13,16,19-octaazabicyclo[6.6.6]icosane, Bifunctional P-450/NADPH-P450 reductase, CHLORIDE ION, ... | | Authors: | Panneerselvm, S, Shehzad, A, Bocola, M, Mueller-Dieckmann, J, Schwaneberg, U. | | Deposit date: | 2015-10-12 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic insights into a cobalt (III) sepulchrate based alternative cofactor system of P450 BM3 monooxygenase.
Biochim. Biophys. Acta, 1866, 2018
|
|
7NEV
 
 | | Structure of the hemiacetal complex between the SARS-CoV-2 Main Protease and Leupeptin | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H.M, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashhour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Xavier, P.L, Ullah, N, Andaleeb, H, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Zaitsev-Doyle, J.J, Rogers, C, Gieseler, H, Melo, D, Monteiro, D.C.F, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schluenzen, F, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Sun, X, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2021-02-05 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6YNQ
 
 | | Structure of SARS-CoV-2 Main Protease bound to 2-Methyl-1-tetralone. | | Descriptor: | (2~{S})-2-methyl-3,4-dihydro-2~{H}-naphthalen-1-one, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6YVF
 
 | | Structure of SARS-CoV-2 Main Protease bound to AZD6482. | | Descriptor: | 2-[[(1R)-1-(7-methyl-2-morpholin-4-yl-4-oxidanylidene-pyrido[1,2-a]pyrimidin-9-yl)ethyl]amino]benzoic acid, 3C-like proteinase, CALCIUM ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-04-28 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7ABU
 
 | | Structure of SARS-CoV-2 Main Protease bound to RS102895 | | Descriptor: | 1'-[2-[4-(trifluoromethyl)phenyl]ethyl]spiro[1~{H}-3,1-benzoxazine-4,4'-piperidine]-2-one, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-09-08 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
