3SRF
 
 | | Human M1 pyruvate kinase | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Morgan, H.P, O'Reilly, F, Palmer, R, McNae, I.W, Nowicki, M.W, Wear, M.A, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2011-07-07 | | Release date: | 2012-08-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.845 Å) | | Cite: | Allosetric regulation of M2 pyruvate kinase.
To be Published
|
|
3SRH
 
 | | Human M2 pyruvate kinase | | Descriptor: | PHOSPHATE ION, Pyruvate kinase isozymes M1/M2 | | Authors: | Morgan, H.P, O'Reilly, F, Palmer, R, McNae, I.W, Nowicki, M.W, Wear, M.A, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2011-07-07 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Allosetric regulation of M2 pyruvate kinase.
To be Published
|
|
3SRD
 
 | | Human M2 pyruvate kinase in complex with fructose 1-6 bisphosphate and Oxalate. | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Morgan, H.P, O'Reilly, F, Palmer, R, McNae, I.W, Nowicki, M.W, Wear, M.A, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2011-07-07 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Allosetric regulation of M2 pyruvate kinase.
To be Published
|
|
1RPH
 
 | |
1RPG
 
 | | STRUCTURES OF RNASE A COMPLEXED WITH 3'-CMP AND D(CPA): ACTIVE SITE CONFORMATION AND CONSERVED WATER MOLECULES | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2'-DEOXYCYTIDINE-2'-DEOXYADENOSINE-3',5'-MONOPHOSPHATE, RIBONUCLEASE A | | Authors: | Zegers, I, Wyns, L, Palmer, R. | | Deposit date: | 1994-08-29 | | Release date: | 1994-12-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The structures of RNase A complexed with 3'-CMP and d(CpA): active site conformation and conserved water molecules.
Protein Sci., 3, 1994
|
|
1RPF
 
 | |
7SP3
 
 | | E. coli RppH bound to Ap4A | | Descriptor: | BIS(ADENOSINE)-5'-TETRAPHOSPHATE, CHLORIDE ION, FLUORIDE ION, ... | | Authors: | Serganov, A.A, Vasilyev, N, Nuthanakanti, A. | | Deposit date: | 2021-11-02 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A distinct RNA recognition mechanism governs Np 4 decapping by RppH.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1RGA
 
 | |
6D1V
 
 | |
6D13
 
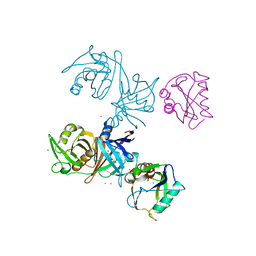 | | Crystal structure of E.coli RppH-DapF complex | | Descriptor: | CHLORIDE ION, Diaminopimelate epimerase, IODIDE ION, ... | | Authors: | Gao, A, Serganov, A. | | Deposit date: | 2018-04-11 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structural and kinetic insights into stimulation of RppH-dependent RNA degradation by the metabolic enzyme DapF.
Nucleic Acids Res., 46, 2018
|
|
6D1Q
 
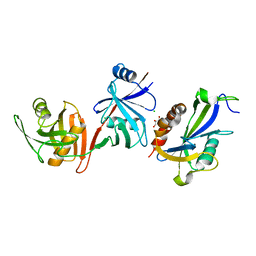 | | Crystal structure of E. coli RppH-DapF complex, monomer | | Descriptor: | CHLORIDE ION, Diaminopimelate epimerase, GLYCEROL, ... | | Authors: | Gao, A, Serganov, A. | | Deposit date: | 2018-04-12 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and kinetic insights into stimulation of RppH-dependent RNA degradation by the metabolic enzyme DapF.
Nucleic Acids Res., 46, 2018
|
|
