4W7U
 
 | |
4W7W
 
 | |
4W7V
 
 | |
5HOS
 
 | |
5HPC
 
 | |
5HNN
 
 | |
2L8A
 
 | | Structure of a novel CBM3 lacking the calcium-binding site | | Descriptor: | Endoglucanase | | Authors: | Paiva, J.H, Meza, A.N, Sforca, M.L, Navarro, R.Z, Neves, J.L, Santos, C.R, Murakami, M.T, Zeri, A.C. | | Deposit date: | 2011-01-07 | | Release date: | 2011-12-21 | | Last modified: | 2011-12-28 | | Method: | SOLUTION NMR | | Cite: | Dissecting structure-function-stability relationships of a thermostable GH5-CBM3 cellulase from Bacillus subtilis 168.
Biochem.J., 441, 2012
|
|
4M1R
 
 | | Structure of a novel cellulase 5 from a sugarcane soil metagenomic library | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cellulase 5 | | Authors: | Paiva, J.H, Alvarez, T.M, Cairo, J.P, Paixao, D.A, Almeida, R.A, Tonoli, C.C.C, Ruiz, D.M, Ruller, R, Santos, C.R, Squina, F.M, Murakami, M.T. | | Deposit date: | 2013-08-03 | | Release date: | 2014-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and function of a novel cellulase 5 from sugarcane soil metagenome.
Plos One, 8, 2013
|
|
6UFV
 
 | |
6UFW
 
 | |
3PZU
 
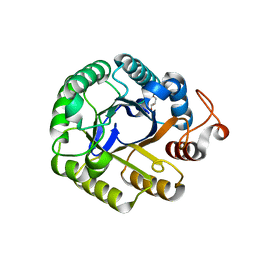 | | P212121 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 | | Descriptor: | Endoglucanase, GLYCEROL | | Authors: | Santos, C.R, Paiva, J.H, Akao, P.K, Meza, A.N, Silva, J.C, Squina, F.M, Ward, R.J, Ruller, R, Murakami, M.T. | | Deposit date: | 2010-12-14 | | Release date: | 2011-09-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dissecting structure-function-stability relationships of a thermostable GH5-CBM3 cellulase from Bacillus subtilis 168.
Biochem.J., 441, 2012
|
|
3PZT
 
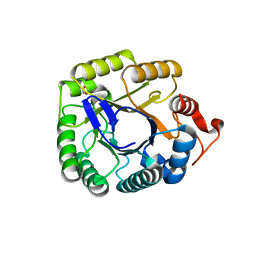 | | Structure of the endo-1,4-beta-glucanase from Bacillus subtilis 168 with manganese(II) ion | | Descriptor: | Endoglucanase, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Santos, C.R, Paiva, J.H, Akao, P.K, Meza, A.N, Silva, J.C, Squina, F.M, Ward, R.J, Ruller, R, Murakami, M.T. | | Deposit date: | 2010-12-14 | | Release date: | 2011-09-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Dissecting structure-function-stability relationships of a thermostable GH5-CBM3 cellulase from Bacillus subtilis 168.
Biochem.J., 441, 2012
|
|
3PZV
 
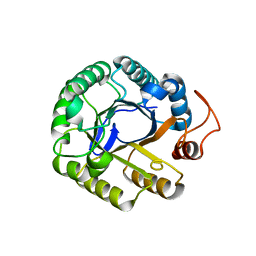 | | C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 | | Descriptor: | Endoglucanase | | Authors: | Santos, C.R, Paiva, J.H, Akao, P.K, Meza, A.N, Silva, J.C, Squina, F.M, Ward, R.J, Ruller, R, Murakami, M.T. | | Deposit date: | 2010-12-14 | | Release date: | 2011-09-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.867 Å) | | Cite: | Dissecting structure-function-stability relationships of a thermostable GH5-CBM3 cellulase from Bacillus subtilis 168.
Biochem.J., 441, 2012
|
|
3PZM
 
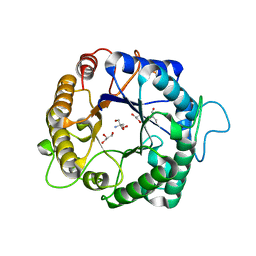 | | Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with three glycerol molecules | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Mannan endo-1,4-beta-mannosidase. Glycosyl Hydrolase family 5 | | Authors: | Santos, C.R, Meza, A.N, Paiva, J.H, Silva, J.C, Ruller, R, Prade, R.A, Squina, F.M, Murakami, M.T. | | Deposit date: | 2010-12-14 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural characterization of a novel hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1
To be Published
|
|
3PZ9
 
 | | Native structure of endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 | | Descriptor: | Mannan endo-1,4-beta-mannosidase. Glycosyl Hydrolase family 5 | | Authors: | Santos, C.R, Meza, A.N, Paiva, J.H, Silva, J.C, Ruller, R, Prade, R.A, Squina, F.M, Murakami, M.T. | | Deposit date: | 2010-12-14 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural characterization of a novel hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1
To be Published
|
|
3PZO
 
 | | Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 in complex with three maltose molecules | | Descriptor: | GLYCEROL, Mannan endo-1,4-beta-mannosidase. Glycosyl Hydrolase family 5, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Santos, C.R, Meza, A.N, Paiva, J.H, Silva, J.C, Ruller, R, Prade, R.A, Squina, F.M, Murakami, M.T. | | Deposit date: | 2010-12-14 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural characterization of a novel hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1
To be Published
|
|
3PZQ
 
 | | Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with maltose and glycerol | | Descriptor: | Mannan endo-1,4-beta-mannosidase. Glycosyl Hydrolase family 5, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Santos, C.R, Meza, A.N, Paiva, J.H, Silva, J.C, Ruller, R, Prade, R.A, Squina, F.M, Murakami, M.T. | | Deposit date: | 2010-12-14 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural characterization of a novel hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1
To be Published
|
|
3PZI
 
 | | Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 in complex with beta-D-glucose | | Descriptor: | Mannan endo-1,4-beta-mannosidase. Glycosyl Hydrolase family 5, beta-D-glucopyranose | | Authors: | Santos, C.R, Meza, A.N, Paiva, J.H, Silva, J.C, Ruller, R, Prade, R.A, Squina, F.M, Murakami, M.T. | | Deposit date: | 2010-12-14 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural characterization of a novel hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1
To be Published
|
|
3PZN
 
 | | Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with citrate and glycerol | | Descriptor: | CITRIC ACID, GLYCEROL, Mannan endo-1,4-beta-mannosidase. Glycosyl Hydrolase family 5 | | Authors: | Santos, C.R, Meza, A.N, Paiva, J.H, Silva, J.C, Ruller, R, Prade, R.A, Squina, F.M, Murakami, M.T. | | Deposit date: | 2010-12-14 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural characterization of a novel hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1
To be Published
|
|
3PZG
 
 | | I222 crystal form of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Mannan endo-1,4-beta-mannosidase. Glycosyl Hydrolase family 5, ... | | Authors: | Santos, C.R, Meza, A.N, Paiva, J.H, Silva, J.C, Ruller, R, Prade, R.A, Squina, F.M, Murakami, M.T. | | Deposit date: | 2010-12-14 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural characterization of a novel hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1
To be Published
|
|
