1AUP
 
 | | GLUTAMATE DEHYDROGENASE | | Descriptor: | NAD-SPECIFIC GLUTAMATE DEHYDROGENASE | | Authors: | Baker, P.J, Waugh, M.L, Stillman, T.J, Turnbull, A.P, Rice, D.W. | | Deposit date: | 1997-09-01 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Determinants of substrate specificity in the superfamily of amino acid dehydrogenases.
Biochemistry, 36, 1997
|
|
1BGV
 
 | | GLUTAMATE DEHYDROGENASE | | Descriptor: | GLUTAMATE DEHYDROGENASE, GLUTAMIC ACID | | Authors: | Stillman, T.J, Baker, P.J, Britton, K.L, Rice, D.W. | | Deposit date: | 1998-06-01 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational flexibility in glutamate dehydrogenase. Role of water in substrate recognition and catalysis.
J.Mol.Biol., 234, 1993
|
|
1B2P
 
 | | NATIVE MANNOSE-SPECIFIC BULB LECTIN FROM SCILLA CAMPANULATA (BLUEBELL) AT 1.7 ANGSTROMS RESOLUTION | | Descriptor: | PROTEIN (LECTIN) | | Authors: | Wood, S.D, Wright, L.M, Reynolds, C.D, Rizkallah, P.J, Allen, A.K, Peumans, W.J, Van Damme, E.J.M. | | Deposit date: | 1998-11-30 | | Release date: | 1999-07-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the native (unligated) mannose-specific bulb lectin from Scilla campanulata (bluebell) at 1.7 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1BDX
 
 | | E. COLI DNA HELICASE RUVA WITH BOUND DNA HOLLIDAY JUNCTION, ALPHA CARBONS AND PHOSPHATE ATOMS ONLY | | Descriptor: | DNA (5'-D(P*GP*CP*AP*TP*GP*CP*AP*TP*AP*TP*GP*CP*AP*TP*GP*C)-3'), HOLLIDAY JUNCTION DNA HELICASE RUVA | | Authors: | Hargreaves, D, Rice, D.W, Sedelnikova, S.E, Artymiuk, P.J, Lloyd, R.G, Rafferty, J.B. | | Deposit date: | 1998-05-11 | | Release date: | 1999-11-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Crystal structure of E.coli RuvA with bound DNA Holliday junction at 6 A resolution.
Nat.Struct.Biol., 5, 1998
|
|
1CDR
 
 | | STRUCTURE OF A SOLUBLE, GLYCOSYLATED FORM OF THE HUMAN COMPLEMENT REGULATORY PROTEIN CD59 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CD59 | | Authors: | Fletcher, C.M, Harrison, R.A, Lachmann, P.J, Neuhaus, D. | | Deposit date: | 1994-06-01 | | Release date: | 1994-09-30 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Structure of a soluble, glycosylated form of the human complement regulatory protein CD59.
Structure, 2, 1994
|
|
1CDS
 
 | | STRUCTURE OF A SOLUBLE, GLYCOSYLATED FORM OF THE HUMAN COMPLEMENT REGULATORY PROTEIN CD59 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD59 | | Authors: | Fletcher, C.M, Harrison, R.A, Lachmann, P.J, Neuhaus, D. | | Deposit date: | 1994-06-01 | | Release date: | 1994-09-30 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Structure of a soluble, glycosylated form of the human complement regulatory protein CD59.
Structure, 2, 1994
|
|
1A14
 
 | | COMPLEX BETWEEN NC10 ANTI-INFLUENZA VIRUS NEURAMINIDASE SINGLE CHAIN ANTIBODY WITH A 5 RESIDUE LINKER AND INFLUENZA VIRUS NEURAMINIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NC10 FV (HEAVY CHAIN), ... | | Authors: | Malby, R.L, Mccoy, A.J, Kortt, A.A, Hudson, P.J, Colman, P.M. | | Deposit date: | 1997-12-21 | | Release date: | 1998-05-13 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structures of single-chain Fv-neuraminidase complexes.
J.Mol.Biol., 279, 1998
|
|
1B4T
 
 | | H48C YEAST CU(II)/ZN SUPEROXIDE DISMUTASE ROOM TEMPERATURE (298K) STRUCTURE | | Descriptor: | CHLORIDE ION, COPPER (II) ION, PROTEIN (CU/ZN SUPEROXIDE DISMUTASE), ... | | Authors: | Hart, P.J, Balbirnie, M.M, Ogihara, N.L, Nersissian, A.M, Weiss, M.S, Valentine, J.S, Eisenberg, D. | | Deposit date: | 1998-12-23 | | Release date: | 1999-12-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
1B4L
 
 | | 15 ATMOSPHERE OXYGEN YEAST CU/ZN SUPEROXIDE DISMUTASE ROOM TEMPERATURE (298K) STRUCTURE | | Descriptor: | COPPER (II) ION, PROTEIN (CU/ZN SUPEROXIDE DISMUTASE), ZINC ION | | Authors: | Hart, P.J, Balbirnie, M.M, Ogihara, N.L, Nersissian, A.M, Weiss, M.S, Valentine, J.S, Eisenberg, D. | | Deposit date: | 1998-12-22 | | Release date: | 1999-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
1BLX
 
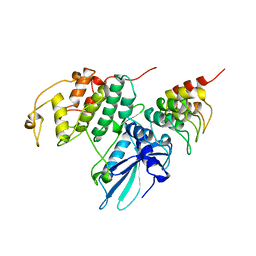 | | P19INK4D/CDK6 COMPLEX | | Descriptor: | CALCIUM ION, CYCLIN-DEPENDENT KINASE 6, P19INK4D | | Authors: | Brotherton, D.H, Dhanaraj, V, Wick, S, Brizuela, L, Domaille, P.J, Volyanik, E, Xu, X, Parisini, E, Smith, B.O, Archer, S.J, Serrano, M, Brenner, S.L, Blundell, T.L, Laue, E.D. | | Deposit date: | 1998-07-21 | | Release date: | 1999-06-01 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the complex of the cyclin D-dependent kinase Cdk6 bound to the cell-cycle inhibitor p19INK4d.
Nature, 395, 1998
|
|
8TIM
 
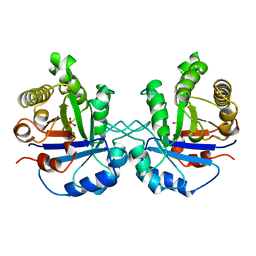 | |
8U7I
 
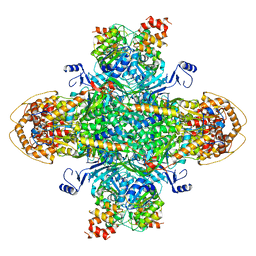 | | Structure of the phage immune evasion protein Gad1 bound to the Gabija GajAB complex | | Descriptor: | Endonuclease GajA, Gabija Anti-Defense 1, Gabija protein GajB | | Authors: | Antine, S.P, Johnson, A.G, Mooney, S.E, Mayer, M.L, Kranzsuch, P.J. | | Deposit date: | 2023-09-15 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structural basis of Gabija anti-phage defence and viral immune evasion.
Nature, 625, 2024
|
|
8QQK
 
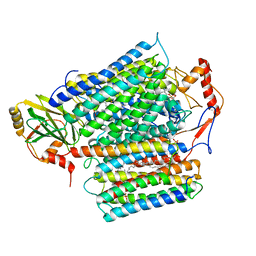 | | Cryo-EM structure of E. coli cytochrome bo3 quinol oxidase assembled in peptidiscs | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, ... | | Authors: | Gao, Y, Zhang, Y, Hakke, S, Peters, P.J, Ravelli, R.B.G. | | Deposit date: | 2023-10-05 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of cytochrome bo 3 quinol oxidase assembled in peptidiscs reveals an "open" conformation for potential ubiquinone-8 release.
Biochim Biophys Acta Bioenerg, 2024
|
|
1AF5
 
 | | GROUP I MOBILE INTRON ENDONUCLEASE | | Descriptor: | I-CREI | | Authors: | Heath, P.J, Stephens, K.M, Monnat Junior, R.J, Stoddard, B.L. | | Deposit date: | 1997-03-21 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of I-Crel, a group I intron-encoded homing endonuclease.
Nat.Struct.Biol., 4, 1997
|
|
1A2W
 
 | | CRYSTAL STRUCTURE OF A 3D DOMAIN-SWAPPED DIMER OF BOVINE PANCREATIC RIBONUCLEASE A | | Descriptor: | CHLORIDE ION, RIBONUCLEASE A, SULFATE ION | | Authors: | Liu, Y, Hart, P.J, Schlunegger, M.P, Eisenberg, D.S. | | Deposit date: | 1998-01-12 | | Release date: | 1998-04-29 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a 3D domain-swapped dimer of RNase A at a 2.1-A resolution.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1AA5
 
 | | VANCOMYCIN | | Descriptor: | ACETIC ACID, CHLORIDE ION, VANCOMYCIN, ... | | Authors: | Loll, P.J, Bevivino, A.E, Korty, B.D, Axelsen, P.H. | | Deposit date: | 1997-01-23 | | Release date: | 1997-08-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Simultaneous Recognition of a Carboxylate-Containing Ligand and an Intramolecular Surrogate Ligand in the Crystal Structure of an Asymmetric Vancomycin Dimer.
J.Am.Chem.Soc., 119, 1997
|
|
1A3X
 
 | | PYRUVATE KINASE FROM SACCHAROMYCES CEREVISIAE COMPLEXED WITH PG, MN2+ AND K+ | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, MANGANESE (II) ION, POTASSIUM ION, ... | | Authors: | Jurica, M.S, Mesecar, A, Heath, P.J, Shi, W, Nowak, T, Stoddard, B.L. | | Deposit date: | 1998-01-26 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The allosteric regulation of pyruvate kinase by fructose-1,6-bisphosphate.
Structure, 6, 1998
|
|
1A3W
 
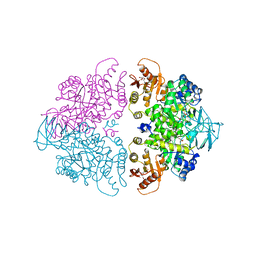 | | PYRUVATE KINASE FROM SACCHAROMYCES CEREVISIAE COMPLEXED WITH FBP, PG, MN2+ AND K+ | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 2-PHOSPHOGLYCOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Jurica, M.S, Mesecar, A, Heath, P.J, Shi, W, Nowak, T, Stoddard, B.L. | | Deposit date: | 1998-01-26 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The allosteric regulation of pyruvate kinase by fructose-1,6-bisphosphate.
Structure, 6, 1998
|
|
1A6Z
 
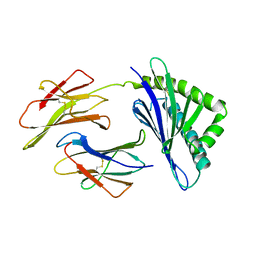 | | HFE (HUMAN) HEMOCHROMATOSIS PROTEIN | | Descriptor: | BETA-2-MICROGLOBULIN, HFE | | Authors: | Lebron, J.A, Bennett, M.J, Vaughn, D.E, Chirino, A.J, Snow, P.M, Mintier, G.A, Feder, J.N, Bjorkman, P.J. | | Deposit date: | 1998-03-04 | | Release date: | 1999-03-23 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the hemochromatosis protein HFE and characterization of its interaction with transferrin receptor.
Cell(Cambridge,Mass.), 93, 1998
|
|
1ABZ
 
 | | ALPHA-T-ALPHA, A DE NOVO DESIGNED PEPTIDE, NMR, 23 STRUCTURES | | Descriptor: | ALPHA-T-ALPHA | | Authors: | Fezoui, Y, Connolly, P.J, Osterhout, J.J. | | Deposit date: | 1997-01-31 | | Release date: | 1998-02-04 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of alpha t alpha, a helical hairpin peptide of de novo design.
Protein Sci., 6, 1997
|
|
1AEW
 
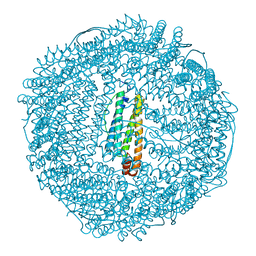 | | L-CHAIN HORSE APOFERRITIN | | Descriptor: | CADMIUM ION, FERRITIN | | Authors: | Hempstead, P.D, Yewdall, S.J, Lawson, D.M, Harrison, P.M, Artymiuk, P.J. | | Deposit date: | 1997-02-26 | | Release date: | 1997-09-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Comparison of the three-dimensional structures of recombinant human H and horse L ferritins at high resolution.
J.Mol.Biol., 268, 1997
|
|
1A68
 
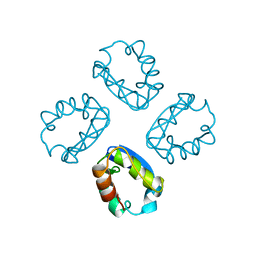 | |
1AZV
 
 | | FAMILIAL ALS MUTANT G37R CUZNSOD (HUMAN) | | Descriptor: | COPPER (II) ION, COPPER/ZINC SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Hart, P.J, Liu, H, Pellegrini, M, Nersissian, A.M, Gralla, E.B, Valentine, J.S, Eisenberg, D. | | Deposit date: | 1997-11-21 | | Release date: | 1998-02-25 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Subunit asymmetry in the three-dimensional structure of a human CuZnSOD mutant found in familial amyotrophic lateral sclerosis.
Protein Sci., 7, 1998
|
|
1AP7
 
 | | P19-INK4D FROM MOUSE, NMR, 20 STRUCTURES | | Descriptor: | P19-INK4D | | Authors: | Archer, S.J, Luh, F.Y, Domaille, P.J, Smith, B.O, Laue, E.D. | | Deposit date: | 1997-07-25 | | Release date: | 1998-09-16 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structure of the cyclin-dependent kinase inhibitor p19Ink4d.
Nature, 389, 1997
|
|
1BHT
 
 | | NK1 FRAGMENT OF HUMAN HEPATOCYTE GROWTH FACTOR | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEPATOCYTE GROWTH FACTOR, SULFATE ION | | Authors: | Ultsch, M.H, Lokker, N.A, Godowski, P.J, De Vos, A.M. | | Deposit date: | 1998-06-10 | | Release date: | 1998-11-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the NK1 fragment of human hepatocyte growth factor at 2.0 A resolution.
Structure, 6, 1998
|
|
