1C9U
 
 | | CRYSTAL STRUCTURE OF THE SOLUBLE QUINOPROTEIN GLUCOSE DEHYDROGENASE IN COMPLEX WITH PQQ | | Descriptor: | CALCIUM ION, GLYCEROL, PYRROLOQUINOLINE QUINONE, ... | | Authors: | Oubrie, A, Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 1999-08-03 | | Release date: | 2000-02-04 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and mechanism of soluble quinoprotein glucose dehydrogenase.
EMBO J., 18, 1999
|
|
1CRU
 
 | | SOLUBLE QUINOPROTEIN GLUCOSE DEHYDROGENASE FROM ACINETOBACTER CALCOACETICUS IN COMPLEX WITH PQQ AND METHYLHYDRAZINE | | Descriptor: | CALCIUM ION, GLYCEROL, METHYLHYDRAZINE, ... | | Authors: | Oubrie, A, Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 1999-08-16 | | Release date: | 2000-03-01 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Active-site structure of the soluble quinoprotein glucose dehydrogenase complexed with methylhydrazine: a covalent cofactor-inhibitor complex.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1QBI
 
 | | SOLUBLE QUINOPROTEIN GLUCOSE DEHYDROGENASE FROM ACINETOBACTER CALCOACETICUS | | Descriptor: | CALCIUM ION, GLYCEROL, PLATINUM (II) ION, ... | | Authors: | Oubrie, A, Rozeboom, H.J, Kalk, K.H, Duine, J.A, Dijkstra, B.W. | | Deposit date: | 1999-04-22 | | Release date: | 2000-02-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The 1.7 A crystal structure of the apo form of the soluble quinoprotein glucose dehydrogenase from Acinetobacter calcoaceticus reveals a novel internal conserved sequence repeat.
J.Mol.Biol., 289, 1999
|
|
1CQ1
 
 | | Soluble Quinoprotein Glucose Dehydrogenase from Acinetobacter Calcoaceticus in Complex with PQQH2 and Glucose | | Descriptor: | CALCIUM ION, PYRROLOQUINOLINE QUINONE, SOLUBLE QUINOPROTEIN GLUCOSE DEHYDROGENASE, ... | | Authors: | Oubrie, A, Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 1999-08-04 | | Release date: | 2000-02-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of soluble quinoprotein glucose dehydrogenase.
EMBO J., 18, 1999
|
|
3SHE
 
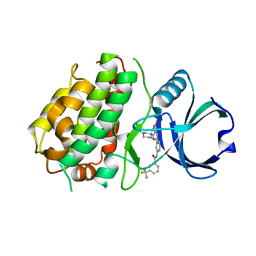 | | Novel ATP-competitive MK2 inhibitors with potent biochemical and cell-based activity throughout the series | | Descriptor: | MAP kinase-activated protein kinase 3, N-{4-[(3S)-4'-oxo-1',4',5',6'-tetrahydrospiro[piperidine-3,7'-pyrrolo[3,2-c]pyridin]-2'-yl]pyridin-2-yl}-3-(trifluoromethyl)benzamide | | Authors: | Oubrie, A, Kazemier, B. | | Deposit date: | 2011-06-16 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Novel ATP competitive MK2 inhibitors with potent biochemical and cell-based activity throughout the series.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
1KB0
 
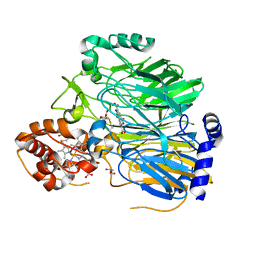 | |
3R2Y
 
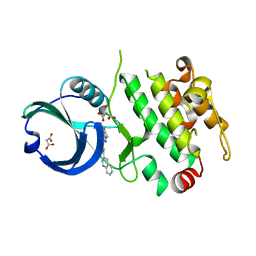 | | MK2 kinase bound to Compound 1 | | Descriptor: | 2-(2-QUINOLIN-3-YLPYRIDIN-4-YL)-1,5,6,7-TETRAHYDRO-4H-PYRROLO[3,2-C]PYRIDIN-4-ONE, MALONATE ION, MAP kinase-activated protein kinase 2 | | Authors: | Oubrie, A, Leonard, P. | | Deposit date: | 2011-03-15 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-based lead identification of ATP-competitive MK2 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3R1N
 
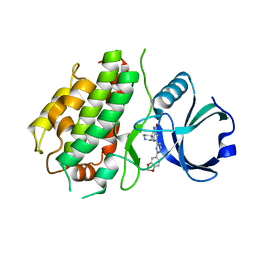 | | MK3 kinase bound to Compound 5b | | Descriptor: | 2'-[2-(1,3-benzodioxol-5-yl)pyrimidin-4-yl]-5',6'-dihydrospiro[piperidine-4,7'-pyrrolo[3,2-c]pyridin]-4'(1'H)-one, MAP kinase-activated protein kinase 3 | | Authors: | Oubrie, A, Kazemier, B. | | Deposit date: | 2011-03-11 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure-based lead identification of ATP-competitive MK2 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3R30
 
 | | MK2 kinase bound to Compound 2 | | Descriptor: | 1-(2-aminoethyl)-3-[2-(quinolin-3-yl)pyridin-4-yl]-1H-pyrazole-5-carboxylic acid, MAP kinase-activated protein kinase 2 | | Authors: | Oubrie, A, Fisher, M. | | Deposit date: | 2011-03-15 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-based lead identification of ATP-competitive MK2 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3R2B
 
 | | MK2 kinase bound to Compound 5b | | Descriptor: | 2'-[2-(1,3-benzodioxol-5-yl)pyrimidin-4-yl]-5',6'-dihydrospiro[piperidine-4,7'-pyrrolo[3,2-c]pyridin]-4'(1'H)-one, MAP kinase-activated protein kinase 2 | | Authors: | Oubrie, A, van Zeeland, M, Versteegh, J. | | Deposit date: | 2011-03-14 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-based lead identification of ATP-competitive MK2 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3DAS
 
 | | Structure of the PQQ-bound form of Aldose Sugar Dehydrogenase (Adh) from Streptomyces coelicolor | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PYRROLOQUINOLINE QUINONE, ... | | Authors: | Southall, S.M, Doel, J.J, Oubrie, A, Richardson, D.J. | | Deposit date: | 2008-05-30 | | Release date: | 2009-06-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Enzymatic Characterization of a Thermostable PQQ-dependent Soluble Aldose Sugar Dehydrogenase
To be Published
|
|
1SU9
 
 | |
1ST9
 
 | | Crystal Structure of a Soluble Domain of ResA in the Oxidised Form | | Descriptor: | 1,2-ETHANEDIOL, Thiol-disulfide oxidoreductase resA | | Authors: | Crow, A, Acheson, R.M, Le Brun, N.E, Oubrie, A. | | Deposit date: | 2004-03-25 | | Release date: | 2004-05-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis of Redox-coupled Protein Substrate Selection by the Cytochrome c Biosynthesis Protein ResA.
J.Biol.Chem., 279, 2004
|
|
2H1B
 
 | | ResA E80Q | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Thiol-disulfide oxidoreductase resA | | Authors: | Lewin, A, Crow, A, Oubrie, A, Le Brun, N.E. | | Deposit date: | 2006-05-16 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular Basis for Specificity of the Extracytoplasmic Thioredoxin ResA.
J.Biol.Chem., 281, 2006
|
|
2H19
 
 | | Crystal Structure of ResA Cys77Ala Variant | | Descriptor: | 1,2-ETHANEDIOL, Thiol-disulfide oxidoreductase resA | | Authors: | Lewin, A, Crow, A, Oubrie, A, Le Brun, N.E. | | Deposit date: | 2006-05-16 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Basis for Specificity of the Extracytoplasmic Thioredoxin ResA.
J.Biol.Chem., 281, 2006
|
|
2H1G
 
 | | ResA C74A/C77A | | Descriptor: | Thiol-disulfide oxidoreductase resA | | Authors: | Lewin, A, Crow, A, Oubrie, A, Le Brun, N.E. | | Deposit date: | 2006-05-16 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular Basis for Specificity of the Extracytoplasmic Thioredoxin ResA.
J.Biol.Chem., 281, 2006
|
|
2H1D
 
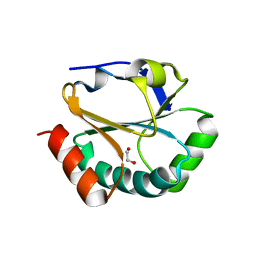 | | ResA pH 9.25 | | Descriptor: | 1,2-ETHANEDIOL, Thiol-disulfide oxidoreductase resA | | Authors: | Lewin, A, Crow, A, Oubrie, A, Le Brun, N.E. | | Deposit date: | 2006-05-16 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Basis for Specificity of the Extracytoplasmic Thioredoxin ResA.
J.Biol.Chem., 281, 2006
|
|
2H1A
 
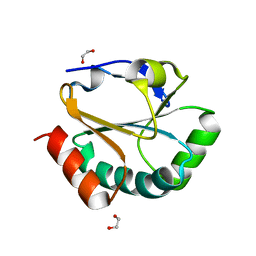 | | ResA C74A Variant | | Descriptor: | 1,2-ETHANEDIOL, Thiol-disulfide oxidoreductase resA | | Authors: | Lewin, A, Crow, A, Oubrie, A, Le Brun, N.E. | | Deposit date: | 2006-05-16 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Basis for Specificity of the Extracytoplasmic Thioredoxin ResA.
J.Biol.Chem., 281, 2006
|
|
3SOS
 
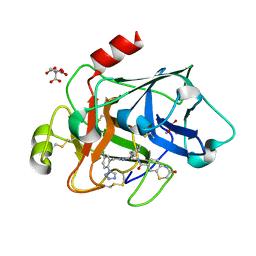 | | Benzothiazinone inhibitor in complex with FXIa | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CITRIC ACID, Coagulation factor XI, ... | | Authors: | Fradera, X, Kazemier, B, Oubrie, A. | | Deposit date: | 2011-06-30 | | Release date: | 2012-04-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | High-resolution crystal structures of factor XIa coagulation factor in complex with nonbasic high-affinity synthetic inhibitors.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3SOR
 
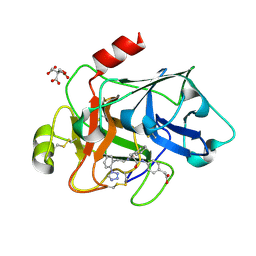 | | Factor XIa in complex with a clorophenyl-tetrazole inhibitor | | Descriptor: | CITRIC ACID, Coagulation factor XI, {4-[(N-{3-[5-chloro-2-(1H-tetrazol-1-yl)phenyl]propanoyl}-L-phenylalanyl)amino]phenyl}acetic acid | | Authors: | Kazemier, B, Oubrie, A. | | Deposit date: | 2011-06-30 | | Release date: | 2012-04-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution crystal structures of factor XIa coagulation factor in complex with nonbasic high-affinity synthetic inhibitors.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
2G8S
 
 | | Crystal structure of the soluble Aldose sugar dehydrogenase (Asd) from Escherichia coli in the apo-form | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glucose/sorbosone dehydrogenases, ... | | Authors: | Southall, S.M, Doel, J.J, Richardson, D.J, Oubrie, A. | | Deposit date: | 2006-03-03 | | Release date: | 2006-08-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Soluble Aldose Sugar Dehydrogenase from Escherichia coli: A HIGHLY EXPOSED ACTIVE SITE CONFERRING BROAD SUBSTRATE SPECIFICITY.
J.Biol.Chem., 281, 2006
|
|
3ZSH
 
 | | X-ray structure of p38alpha bound to SCIO-469 | | Descriptor: | 2-(6-chloro-5-{[(2R,5S)-4-(4-fluorobenzyl)-2,5-dimethylpiperazin-1-yl]carbonyl}-1-methyl-1H-indol-3-yl)-N,N-dimethyl-2-oxoacetamide, MITOGEN-ACTIVATED PROTEIN KINASE 14, octyl beta-D-glucopyranoside | | Authors: | Azevedo, R, van Zeeland, M, Raaijmakers, H, Kazemier, B, Oubrie, A. | | Deposit date: | 2011-06-28 | | Release date: | 2012-06-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray structure of p38 alpha bound to TAK-715: comparison with three classic inhibitors.
Acta Crystallogr. D Biol. Crystallogr., 68, 2012
|
|
3ZRA
 
 | | Structural basis for agonism and antagonism for a set of chemically related progesterone receptor modulators | | Descriptor: | N-{(1R)-1-[4-(2-CHLORO-5-FLUOROPYRIDIN-3-YL)PHENYL]ETHYL}-3,5-DIMETHYLISOXAZOLE-4-SULFONAMIDE, PROGESTERONE RECEPTOR, SULFATE ION | | Authors: | Lusher, S.J, Raaijmakers, H.C.A, Vu-Pham, D, Dechering, K, Wai Lam, T, Brown, A.R, Hamilton, N.M, Nimz, O, Azevedo, R, McGuire, R, Oubrie, A, de Vlieg, J. | | Deposit date: | 2011-06-15 | | Release date: | 2011-08-17 | | Last modified: | 2017-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for agonism and antagonism for a set of chemically related progesterone receptor modulators.
J. Biol. Chem., 286, 2011
|
|
3ZSG
 
 | | X-ray structure of p38alpha bound to TAK-715 | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE 14, TAK-715, octyl beta-D-glucopyranoside | | Authors: | Azevedo, R, van Zeeland, M, Raaijmakers, H, Kazemier, B, Oubrie, A. | | Deposit date: | 2011-06-28 | | Release date: | 2012-06-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | X-ray structure of p38 alpha bound to TAK-715: comparison with three classic inhibitors.
Acta Crystallogr. D Biol. Crystallogr., 68, 2012
|
|
3ZRB
 
 | | Structural basis for agonism and antagonism for a set of chemically related progesterone receptor modulators | | Descriptor: | (R)-N-[1-[4-(3,5-DIMETHYLISOXAZOL-4-YL)PHENYL]ETHYL]-3,5-DIMETHYLISOXAZOLE-4-SULFONAMIDE, 2-CHLORO-N-[[4-(3,5-DIMETHYLISOXAZOL-4-YL)PHENYL]METHYL]-1,4-DIMETHYL-1H-PYRAZOLE-4-SULFONAMIDE, GLYCEROL, ... | | Authors: | Lusher, S.J, Raaijmakers, H.C.A, Vu-Pham, D, Dechering, K, Wai Lam, T, Brown, A.R, Hamilton, N.M, Nimz, O, Azevedo, R, Mcguire, R, Oubrie, A, de Vlieg, J. | | Deposit date: | 2011-06-15 | | Release date: | 2011-08-17 | | Last modified: | 2017-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for agonism and antagonism for a set of chemically related progesterone receptor modulators.
J. Biol. Chem., 286, 2011
|
|
