6A9E
 
 | | Crystal structure of the N-terminal domain of Atg2 | | Descriptor: | Endolysin,Autophagy-related protein 2 | | Authors: | Osawa, T, Noda, N.N. | | Deposit date: | 2018-07-13 | | Release date: | 2019-03-20 | | Last modified: | 2019-04-17 | | Method: | X-RAY DIFFRACTION (3.205 Å) | | Cite: | Atg2 mediates direct lipid transfer between membranes for autophagosome formation.
Nat. Struct. Mol. Biol., 26, 2019
|
|
6A9J
 
 | | Crystal structure of the PE-bound N-terminal domain of Atg2 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Endolysin,Autophagy-related protein 2 | | Authors: | Osawa, T, Noda, N.N. | | Deposit date: | 2018-07-13 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Atg2 mediates direct lipid transfer between membranes for autophagosome formation.
Nat. Struct. Mol. Biol., 26, 2019
|
|
8GZP
 
 | | Cryo-EM structure of the NS5-SLA complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Genome polyprotein, MAGNESIUM ION, ... | | Authors: | Osawa, T, Ehara, H, Sekine, S. | | Deposit date: | 2022-09-27 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of dengue virus RNA replicase complexes.
Mol.Cell, 83, 2023
|
|
8GZR
 
 | | Cryo-EM structure of the the NS5-NS3 RNA-elongation complex | | Descriptor: | CYTIDINE-5'-DIPHOSPHATE, Genome polyprotein, MANGANESE (II) ION, ... | | Authors: | Osawa, T, Ehara, H, Sekine, S. | | Deposit date: | 2022-09-27 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of dengue virus RNA replicase complexes.
Mol.Cell, 83, 2023
|
|
8GZQ
 
 | | Cryo-EM structure of the NS5-NS3-SLA complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Genome polyprotein, RNA, ... | | Authors: | Osawa, T, Ehara, H, Sekine, S. | | Deposit date: | 2022-09-27 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of dengue virus RNA replicase complexes.
Mol.Cell, 83, 2023
|
|
2Z87
 
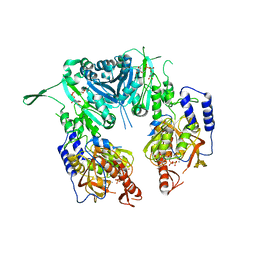 | | Crystal structure of chondroitin polymerase from Escherichia coli strain K4 (K4CP) complexed with UDP-GalNAc and UDP | | Descriptor: | Chondroitin synthase, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Osawa, T, Sugiura, N, Shimada, H, Hirooka, R, Tsuji, A, Kimura, M, Kimata, K, Kakuta, Y. | | Deposit date: | 2007-09-03 | | Release date: | 2008-09-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of chondroitin polymerase from Escherichia coli K4.
Biochem. Biophys. Res. Commun., 378, 2009
|
|
2Z86
 
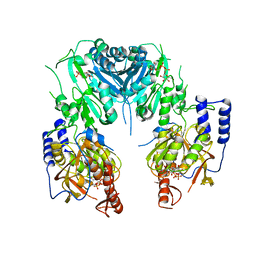 | | Crystal structure of chondroitin polymerase from Escherichia coli strain K4 (K4CP) complexed with UDP-GlcUA and UDP | | Descriptor: | Chondroitin synthase, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Osawa, T, Sugiura, N, Shimada, H, Hirooka, R, Tsuji, A, Kimura, M, Kimata, K, Kakuta, Y. | | Deposit date: | 2007-09-03 | | Release date: | 2008-09-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of chondroitin polymerase from Escherichia coli K4.
Biochem. Biophys. Res. Commun., 378, 2009
|
|
3X1L
 
 | |
3W2W
 
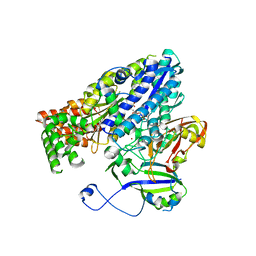 | | Crystal structure of the Cmr2dHD-Cmr3 subcomplex bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR system Cmr subunit Cmr2, CRISPR system Cmr subunit Cmr3, ... | | Authors: | Numata, T, Osawa, T. | | Deposit date: | 2012-12-06 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Cmr2-Cmr3 Subcomplex in the CRISPR-Cas RNA Silencing Effector Complex.
J.Mol.Biol., 425, 2013
|
|
3W2V
 
 | |
3AMU
 
 | |
3AMT
 
 | | Crystal structure of the TiaS-tRNA(Ile2)-ATP complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Putative uncharacterized protein, RNA (78-MER) | | Authors: | Numata, T, Osawa, T. | | Deposit date: | 2010-08-23 | | Release date: | 2011-10-12 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of tRNA agmatinylation essential for AUA codon decoding
Nat.Struct.Mol.Biol., 18, 2011
|
|
3AU7
 
 | |
2ZXI
 
 | | Structure of Aquifex aeolicus GidA in the form II crystal | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, tRNA uridine 5-carboxymethylaminomethyl modification enzyme mnmG | | Authors: | Numata, T, Osawa, T. | | Deposit date: | 2008-12-24 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conserved cysteine residues of GidA are essential for biogenesis of 5-carboxymethylaminomethyluridine at tRNA anticodon
Structure, 17, 2009
|
|
2ZXH
 
 | | Structure of Aquifex aeolicus GidA in the form I crystal | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, tRNA uridine 5-carboxymethylaminomethyl modification enzyme mnmG | | Authors: | Numata, T, Osawa, T. | | Deposit date: | 2008-12-24 | | Release date: | 2009-05-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Conserved cysteine residues of GidA are essential for biogenesis of 5-carboxymethylaminomethyluridine at tRNA anticodon
Structure, 17, 2009
|
|
4QTS
 
 | |
4MNJ
 
 | | Crystal Structure of GH18 Chitinase from Cycad, Cycas revoluta | | Descriptor: | ACETATE ION, Chitinase A | | Authors: | Umemoto, N, Numata, T, Ohnuma, T, Osawa, T, Taira, T, Fukamizo, T. | | Deposit date: | 2013-09-11 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of GH18 Chitinase from Cycad, Cycas revoluta
To be Published
|
|
4MNL
 
 | | Crystal Structure of GH18 Chitinase (G77W/E119Q mutant) from Cycas revoluta in complex with (GlcNAc)4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Chitinase A | | Authors: | Umemoto, N, Numata, T, Ohnuma, T, Osawa, T, Taira, T, Fukamizo, T. | | Deposit date: | 2013-09-11 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of GH18 Chitinase form Cycad, Cycas revoluta
To be Published
|
|
4MNM
 
 | | Crystal Structure of GH18 Chitinase (G77W/E119Q mutant) from Cycas revoluta in complex with (GlcNAc)4 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase A, ... | | Authors: | Umemoto, N, Numata, T, Ohnuma, T, Osawa, T, Taira, T, Fukamizo, T. | | Deposit date: | 2013-09-11 | | Release date: | 2014-09-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of GH18 Chitinase from Cycad, Cycas revoluta
To be Published
|
|
4MNK
 
 | | Crystal Structure of GH18 Chitinase from Cycas revoluta in complex with (GlcNAc)3 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Umemoto, N, Numata, T, Ohnuma, T, Osawa, T, Taira, T, Fukamizo, T. | | Deposit date: | 2013-09-11 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Crystal Structure of GH18 Chitinase from Cycad, Cycas revoluta
To be Published
|
|
3WIJ
 
 | | Crystal structure of a plant class V chitinase mutant from Cycas revoluta in complex with (GlcNAc)3 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Numata, T, Osawa, T, Umemoto, N, Ohnuma, T, Fukamizo, T. | | Deposit date: | 2013-09-13 | | Release date: | 2014-09-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of a plant class V chitinase mutant from Cycas revoluta in complex with (GlcNAc)3
To be Published
|
|
2Z7C
 
 | | Crystal structure of chromatin protein alba from hyperthermophilic archaeon pyrococcus horikoshii | | Descriptor: | ARGININE, DNA/RNA-binding protein Alba | | Authors: | Hada, K, Nakashima, T, Osawa, T, Shimada, H, Kakuta, Y, Kimura, M. | | Deposit date: | 2007-08-17 | | Release date: | 2008-08-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure and functional analysis of an archaeal chromatin protein Alba from the hyperthermophilic archaeon Pyrococcus horikoshii OT3.
Biosci.Biotechnol.Biochem., 72, 2008
|
|
3ALF
 
 | | Crystal Structure of Class V Chitinase from Nicotiana tobaccum | | Descriptor: | 1,2-ETHANEDIOL, Chitinase, class V, ... | | Authors: | Numata, T, Osawa, T, Ohnuma, T, Fukamizo, T. | | Deposit date: | 2010-08-03 | | Release date: | 2011-03-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure and mode of action of a class V chitinase from Nicotiana tabacum
Plant Mol.Biol., 75, 2011
|
|
3ALG
 
 | | Crystal Structure of Class V Chitinase (E115Q mutant) from Nicotiana tobaccum in complex with NAG4 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase, ... | | Authors: | Numata, T, Osawa, T, Ohnuma, T, Fukamizo, T. | | Deposit date: | 2010-08-03 | | Release date: | 2011-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and mode of action of a class V chitinase from Nicotiana tabacum
Plant Mol.Biol., 75, 2011
|
|
3AQU
 
 | | Crystal structure of a class V chitinase from Arabidopsis thaliana | | Descriptor: | At4g19810, CITRATE ANION | | Authors: | Numata, T, Ohnuma, T, Osawa, T, Fukamizo, T. | | Deposit date: | 2010-11-19 | | Release date: | 2011-07-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A class V chitinase from Arabidopsis thaliana: gene responses, enzymatic properties, and crystallographic analysis
Planta, 234, 2011
|
|
