6HCY
 
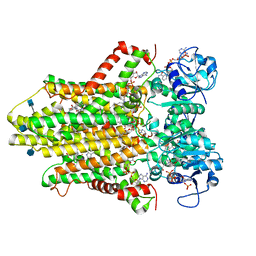 | | human STEAP4 bound to NADP, FAD, heme and Fe(III)-NTA. | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Oosterheert, W, van Bezouwen, L.S, Rodenburg, R.N.P, Forster, F, Mattevi, A, Gros, P. | | Deposit date: | 2018-08-17 | | Release date: | 2018-10-24 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of human STEAP4 reveal mechanism of iron(III) reduction.
Nat Commun, 9, 2018
|
|
6Z1V
 
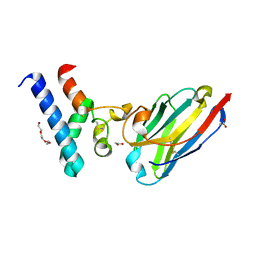 | | Structure of the EC2 domain of CD9 in complex with nanobody 4E8 | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CD9 antigen, ... | | Authors: | Oosterheert, W, Pearce, N.M, Gros, P. | | Deposit date: | 2020-05-14 | | Release date: | 2020-09-23 | | Last modified: | 2020-09-30 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Implications for tetraspanin-enriched microdomain assembly based on structures of CD9 with EWI-F.
Life Sci Alliance, 3, 2020
|
|
6Z20
 
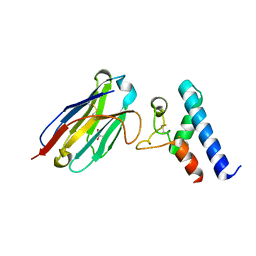 | | Structure of the EC2 domain of CD9 in complex with nanobody 4C8 | | Descriptor: | CD9 antigen, CHLORIDE ION, GLYCEROL, ... | | Authors: | Oosterheert, W, Manshande, J, Pearce, N.M, Lutz, M, Gros, P. | | Deposit date: | 2020-05-14 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Implications for tetraspanin-enriched microdomain assembly based on structures of CD9 with EWI-F.
Life Sci Alliance, 3, 2020
|
|
6HD1
 
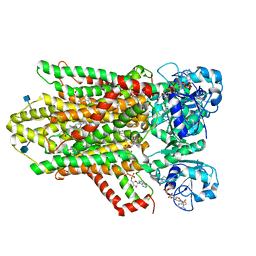 | | human STEAP4 bound to NADPH, FAD and heme. | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Oosterheert, W, van Bezouwen, L.S, Rodenburg, R.N.P, Forster, F, Mattevi, A, Gros, P. | | Deposit date: | 2018-08-17 | | Release date: | 2018-10-24 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of human STEAP4 reveal mechanism of iron(III) reduction.
Nat Commun, 9, 2018
|
|
6Y9B
 
 | |
8OI6
 
 | | Cryo-EM structure of the undecorated barbed end of filamentous beta/gamma actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Oosterheert, W, Blanc, F.E.C, Roy, A, Belyy, A, Hofnagel, O, Hummer, G, Bieling, P, Raunser, S. | | Deposit date: | 2023-03-22 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Molecular mechanisms of inorganic-phosphate release from the core and barbed end of actin filaments.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8OID
 
 | | Cryo-EM structure of ADP-bound, filamentous beta-actin harboring the N111S mutation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Oosterheert, W, Blanc, F.E.C, Roy, A, Belyy, A, Hofnagel, O, Hummer, G, Bieling, P, Raunser, S. | | Deposit date: | 2023-03-22 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Molecular mechanisms of inorganic-phosphate release from the core and barbed end of actin filaments.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8OI8
 
 | | Cryo-EM structure of ADP-bound, filamentous beta-actin harboring the R183W mutation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Oosterheert, W, Blanc, F.E.C, Roy, A, Belyy, A, Hofnagel, O, Hummer, G, Bieling, P, Raunser, S. | | Deposit date: | 2023-03-22 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.28 Å) | | Cite: | Molecular mechanisms of inorganic-phosphate release from the core and barbed end of actin filaments.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8RTY
 
 | | Structure of the F-actin barbed end bound by Cdc12 and profilin (ring complex) at a resolution of 6.3 Angstrom | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Oosterheert, W, Boiero Sanders, M, Funk, J, Prumbaum, D, Raunser, S, Bieling, P. | | Deposit date: | 2024-01-29 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (6.25 Å) | | Cite: | Molecular mechanism of actin filament elongation by formins
Science, 384, 2024
|
|
8RU0
 
 | | Structure of the undecorated barbed end of F-actin. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Oosterheert, W, Boiero Sanders, M, Funk, J, Prumbaum, D, Raunser, S, Bieling, P. | | Deposit date: | 2024-01-29 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Molecular mechanism of actin filament elongation by formins
Science, 384, 2024
|
|
8RU2
 
 | | Structure of the F-actin barbed end bound by formin mDia1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Oosterheert, W, Boiero Sanders, M, Funk, J, Prumbaum, D, Raunser, S, Bieling, P. | | Deposit date: | 2024-01-29 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Molecular mechanism of actin filament elongation by formins
Science, 384, 2024
|
|
8RTT
 
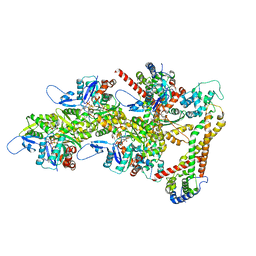 | | Structure of the formin Cdc12 bound to the barbed end of phalloidin-stabilized F-actin. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Oosterheert, W, Boiero Sanders, M, Funk, J, Prumbaum, D, Raunser, S, Bieling, P. | | Deposit date: | 2024-01-29 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Mechanism of formin-mediated processive elongation of actin filaments revealed by cryo-EM
To Be Published
|
|
8RV2
 
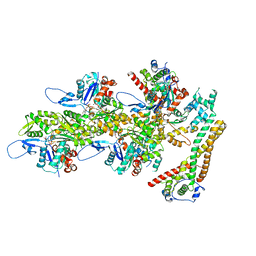 | | Structure of the formin INF2 bound to the barbed end of F-actin. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Oosterheert, W, Boiero Sanders, M, Funk, J, Prumbaum, D, Raunser, S, Bieling, P. | | Deposit date: | 2024-01-31 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Molecular mechanism of actin filament elongation by formins
Science, 384, 2024
|
|
8A2Z
 
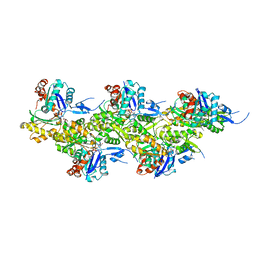 | | Cryo-EM structure of F-actin in the Ca2+-ADP nucleotide state. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Oosterheert, W, Klink, B.U, Belyy, A, Pospich, S, Raunser, S. | | Deposit date: | 2022-06-06 | | Release date: | 2022-08-10 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | Structural basis of actin filament assembly and aging.
Nature, 611, 2022
|
|
8A2Y
 
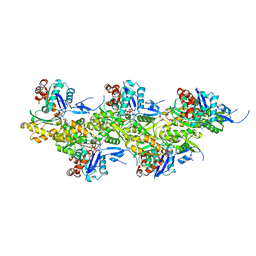 | | Cryo-EM structure of F-actin in the Ca2+-ADP-Pi nucleotide state. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Oosterheert, W, Klink, B.U, Belyy, A, Pospich, S, Raunser, S. | | Deposit date: | 2022-06-06 | | Release date: | 2022-08-10 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | Structural basis of actin filament assembly and aging.
Nature, 611, 2022
|
|
8A2T
 
 | | Cryo-EM structure of F-actin in the Mg2+-ADP nucleotide state. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Oosterheert, W, Klink, B.U, Belyy, A, Pospich, S, Raunser, S. | | Deposit date: | 2022-06-06 | | Release date: | 2022-08-10 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | Structural basis of actin filament assembly and aging.
Nature, 611, 2022
|
|
8A2U
 
 | | Cryo-EM structure of F-actin in the Ca2+-ADP-BeF3- nucleotide state. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Oosterheert, W, Klink, B.U, Belyy, A, Pospich, S, Raunser, S. | | Deposit date: | 2022-06-06 | | Release date: | 2022-08-10 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.21 Å) | | Cite: | Structural basis of actin filament assembly and aging.
Nature, 611, 2022
|
|
8A2S
 
 | | Cryo-EM structure of F-actin in the Mg2+-ADP-Pi nucleotide state. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Oosterheert, W, Klink, B.U, Belyy, A, Pospich, S, Raunser, S. | | Deposit date: | 2022-06-06 | | Release date: | 2022-08-10 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.22 Å) | | Cite: | Structural basis of actin filament assembly and aging.
Nature, 611, 2022
|
|
8A2R
 
 | | Cryo-EM structure of F-actin in the Mg2+-ADP-BeF3- nucleotide state. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Oosterheert, W, Klink, B.U, Belyy, A, Pospich, S, Raunser, S. | | Deposit date: | 2022-06-06 | | Release date: | 2022-08-10 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.17 Å) | | Cite: | Structural basis of actin filament assembly and aging.
Nature, 611, 2022
|
|
6Z1Z
 
 | | Structure of the anti-CD9 nanobody 4C8 | | Descriptor: | Nanobody 4C8 | | Authors: | Neviani, N, Oosterheert, W, Pearce, N.M, Lutz, M, Kroon-Batenburg, L.M.J, Gros, P. | | Deposit date: | 2020-05-14 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Implications for tetraspanin-enriched microdomain assembly based on structures of CD9 with EWI-F.
Life Sci Alliance, 3, 2020
|
|
7B2M
 
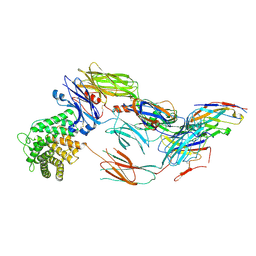 | | Cryo-EM structure of complement C4b in complex with nanobody E3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Anti-C4b nanobody E3, Complement C4 alpha chain, ... | | Authors: | Oosterheert, W, De la O Becerra, K.I, van den Bos, R.M, Gros, P. | | Deposit date: | 2020-11-27 | | Release date: | 2022-03-02 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Multifaceted Activities of Seven Nanobodies against Complement C4b.
J Immunol., 208, 2022
|
|
7B2P
 
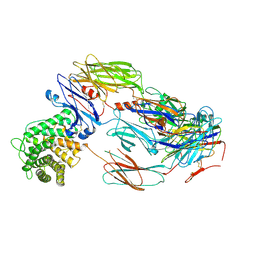 | | Cryo-EM structure of complement C4b in complex with nanobody B5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C4 alpha chain, ... | | Authors: | Oosterheert, W, De la O Becerra, K.I, Gros, P. | | Deposit date: | 2020-11-27 | | Release date: | 2022-03-02 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Multifaceted Activities of Seven Nanobodies against Complement C4b.
J Immunol., 208, 2022
|
|
7B2Q
 
 | | Cryo-EM structure of complement C4b in complex with nanobody B12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Anti-C4b nanobody B12, ... | | Authors: | Oosterheert, W, De la O Becerra, K.I, Gros, P. | | Deposit date: | 2020-11-27 | | Release date: | 2022-03-02 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Multifaceted Activities of Seven Nanobodies against Complement C4b.
J Immunol., 208, 2022
|
|
5SVR
 
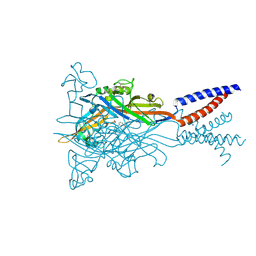 | | Crystal structure of the ATP-gated human P2X3 ion channel bound to competitive antagonist A-317491 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-{[(3-phenoxyphenyl)methyl][(1S)-1,2,3,4-tetrahydronaphthalen-1-yl]carbamoyl}benzene-1,2,4-tricarboxylic acid, MAGNESIUM ION, ... | | Authors: | Mansoor, S.E, Lu, W, Oosterheert, W, Shekhar, M, Tajkhorshid, E, Gouaux, E. | | Deposit date: | 2016-08-07 | | Release date: | 2016-10-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | X-ray structures define human P2X3 receptor gating cycle and antagonist action.
Nature, 538, 2016
|
|
5SVJ
 
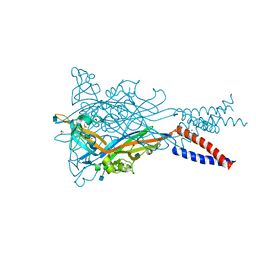 | | Crystal structure of the ATP-gated human P2X3 ion channel in the closed, apo state | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, ... | | Authors: | Mansoor, S.E, Lu, W, Oosterheert, W, Shekhar, M, Tajkhorshid, E, Gouaux, E. | | Deposit date: | 2016-08-06 | | Release date: | 2016-09-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.984 Å) | | Cite: | X-ray structures define human P2X3 receptor gating cycle and antagonist action.
Nature, 538, 2016
|
|
