1UOY
 
 | | The bubble protein from Penicillium brevicompactum Dierckx exudate. | | Descriptor: | BUBBLE PROTEIN | | Authors: | Olsen, J.G, Flensburg, C, Olsen, O, Seibold, M, Bricogne, G, Henriksen, A. | | Deposit date: | 2003-09-26 | | Release date: | 2003-11-04 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Solving the Structure of the Bubble Protein Using the Anomalous Sulfur Signal from Single-Crystal in-House Cu Kalpha Diffraction Data Only
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2VDG
 
 | | Barley Aldose Reductase 1 complex with butanol | | Descriptor: | 1-BUTANOL, ALDOSE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Olsen, J.G, Pedersen, L, Christensen, C.L, Olsen, O, Henriksen, A. | | Deposit date: | 2007-10-08 | | Release date: | 2008-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Barley Aldose Reductase: Structure, Cofactor Binding, and Substrate Recognition in the Aldo/Keto Reductase 4C Family.
Proteins: Struct., Funct., Bioinf., 71, 2008
|
|
2BGQ
 
 | | apo aldose reductase from barley | | Descriptor: | ALDOSE REDUCTASE, SULFATE ION | | Authors: | Olsen, J.G, Pedersen, L, Christensen, C.L, Olsen, O, Henriksen, A. | | Deposit date: | 2005-01-04 | | Release date: | 2006-06-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Barley Aldose Reductase: Structure, Cofactor Binding, and Substrate Recognition in the Aldo/Keto Reductase 4C Family.
Proteins: Struct., Funct., Bioinf., 71, 2008
|
|
2BGS
 
 | | HOLO ALDOSE REDUCTASE FROM BARLEY | | Descriptor: | ALDOSE REDUCTASE, BICARBONATE ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Olsen, J.G, Pedersen, L, Christensen, C.L, Olsen, O, Henriksen, A. | | Deposit date: | 2005-01-05 | | Release date: | 2006-06-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Barley Aldose Reductase: Structure, Cofactor Binding, and Substrate Recognition in the Aldo/Keto Reductase 4C Family.
Proteins: Struct., Funct., Bioinf., 71, 2008
|
|
1KUN
 
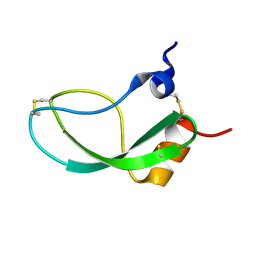 | | SOLUTION STRUCTURE OF THE HUMAN ALPHA3-CHAIN TYPE VI COLLAGEN C-TERMINAL KUNITZ DOMAIN, NMR, 20 STRUCTURES | | Descriptor: | ALPHA3-CHAIN TYPE VI COLLAGEN | | Authors: | Sorensen, M.D, Bjorn, S, Norris, K, Olsen, O, Petersen, L, James, T.L, Led, J.J. | | Deposit date: | 1997-03-04 | | Release date: | 1997-11-12 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of the human alpha3-chain type VI collagen C-terminal Kunitz domain,.
Biochemistry, 36, 1997
|
|
1KNT
 
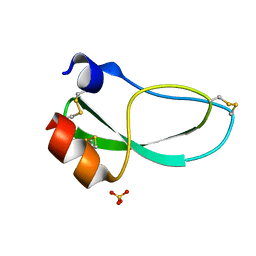 | | THE 1.6 ANGSTROMS STRUCTURE OF THE KUNITZ-TYPE DOMAIN FROM THE ALPHA3 CHAIN OF THE HUMAN TYPE VI COLLAGEN | | Descriptor: | COLLAGEN TYPE VI, SULFATE ION | | Authors: | Arnoux, B, Merigeau, K, Saludjian, P, Norris, F, Norris, K, Bjorn, S, Olsen, O, Petersen, L, Ducruix, A. | | Deposit date: | 1994-08-18 | | Release date: | 1994-11-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1.6 A structure of Kunitz-type domain from the alpha 3 chain of human type VI collagen.
J.Mol.Biol., 246, 1995
|
|
1KJW
 
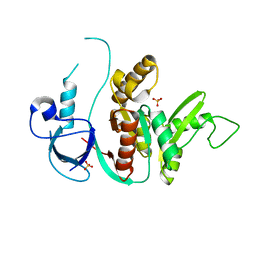 | | SH3-Guanylate Kinase Module from PSD-95 | | Descriptor: | POSTSYNAPTIC DENSITY PROTEIN 95, SULFATE ION | | Authors: | McGee, A.W, Dakoji, S.R, Olsen, O, Bredt, D.S, Lim, W.A, Prehoda, K.E. | | Deposit date: | 2001-12-05 | | Release date: | 2002-01-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the SH3-Guanylate Kinase Module from PSD-95 Suggests a Mechanism for Regulated Assembly of MAGUK Scaffolding Proteins
Mol.Cell, 8, 2001
|
|
1BYH
 
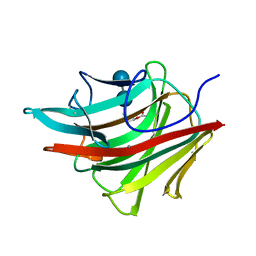 | | MOLECULAR AND ACTIVE-SITE STRUCTURE OF A BACILLUS (1-3,1-4)-BETA-GLUCANASE | | Descriptor: | CALCIUM ION, HYBRID, N-BUTANE, ... | | Authors: | Keitel, T, Heinemann, U. | | Deposit date: | 1992-12-31 | | Release date: | 1993-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular and active-site structure of a Bacillus 1,3-1,4-beta-glucanase.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
4YN0
 
 | | Crystal structure of APP E2 domain in complex with DR6 CRD domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Amyloid beta A4 protein, MAGNESIUM ION, ... | | Authors: | Xu, K, Nikolov, D. | | Deposit date: | 2015-03-08 | | Release date: | 2015-04-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of DR6 in complex with the amyloid precursor protein provides insight into death receptor activation.
Genes Dev., 29, 2015
|
|
1MAC
 
 | | CRYSTAL STRUCTURE AND SITE-DIRECTED MUTAGENESIS OF BACILLUS MACERANS ENDO-1,3-1,4-BETA-GLUCANASE | | Descriptor: | 1,3-1,4-BETA-D-GLUCAN 4-GLUCANOHYDROLASE, CALCIUM ION | | Authors: | Hahn, M, Heinemann, U. | | Deposit date: | 1994-12-22 | | Release date: | 1995-02-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and site-directed mutagenesis of Bacillus macerans endo-1,3-1,4-beta-glucanase.
J.Biol.Chem., 270, 1995
|
|
2XKX
 
 | | Single particle analysis of PSD-95 in negative stain | | Descriptor: | DISKS LARGE HOMOLOG 4 | | Authors: | Fomina, S, Howard, T.D, Sleator, O.K, Golovanova, M, O'Ryan, L, Leyland, M.L, Grossmann, J.G, Collins, R.F, Prince, S.M. | | Deposit date: | 2010-07-15 | | Release date: | 2011-07-20 | | Last modified: | 2019-11-13 | | Method: | ELECTRON MICROSCOPY (22.9 Å), SOLUTION SCATTERING | | Cite: | Self-Directed Assembly and Clustering of the Cytoplasmic Domains of Inwardly Rectifying Kir2.1 Potassium Channels on Association with Psd-95
Biochim.Biophys.Acta, 1808, 2011
|
|
1KTH
 
 | |
