1BMQ
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF INTERLEUKIN-1BETA CONVERTING ENZYME (ICE) WITH A PEPTIDE BASED INHIBITOR, (3S )-N-METHANESULFONYL-3-({1-[N-(2-NAPHTOYL)-L-VALYL]-L-PROLYL }AMINO)-4-OXOBUTANAMIDE | | Descriptor: | (3S)-N-METHANESULFONYL-3-({1-[N-(2-NAPHTOYL)-L-VALYL]-L-PROLYL}AMINO)-4-OXOBUTANAMIDE, PROTEIN (INTERLEUKIN-1 BETA CONVERTASE) | | Authors: | Okamoto, Y, Anan, H, Nakai, E, Morihira, K, Yonetoku, Y, Kurihara, H, Katayama, N, Sakashita, H, Terai, Y, Takeuchi, M, Shibanuma, T, Isomura, Y. | | Deposit date: | 1998-07-24 | | Release date: | 1998-07-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Peptide based interleukin-1 beta converting enzyme (ICE) inhibitors: synthesis, structure activity relationships and crystallographic study of the ICE-inhibitor complex.
Chem.Pharm.Bull., 47, 1999
|
|
3WHN
 
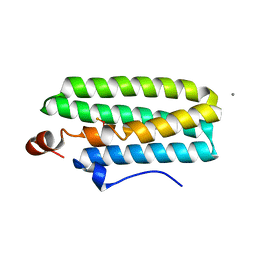 | | Hemerythrin-like domain of DcrH I119H mutant (met) | | Descriptor: | CALCIUM ION, CHLORO DIIRON-OXO MOIETY, Hemerythrin-like domain protein DcrH | | Authors: | Okamoto, Y, Onoda, A, Sugimoto, H, Takano, Y, Hirota, S, Kurtz Jr, D.M, Shiro, Y, Hayashi, T. | | Deposit date: | 2013-08-29 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | H2O2-dependent substrate oxidation by an engineered diiron site in a bacterial hemerythrin.
Chem.Commun.(Camb.), 50, 2014
|
|
3WAQ
 
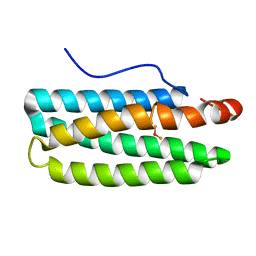 | | Hemerythrin-like domain of DcrH I119E mutant (met) | | Descriptor: | Hemerythrin-like domain protein DcrH, MU-OXO-DIIRON | | Authors: | Okamoto, Y, Onoda, A, Sugimoto, H, Takano, Y, Hirota, S, Kurtz Jr, D.M, Shiro, Y, Hayashi, T. | | Deposit date: | 2013-05-07 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure, exogenous ligand binding, and redox properties of an engineered diiron active site in a bacterial hemerythrin
Inorg.Chem., 52, 2013
|
|
5XJE
 
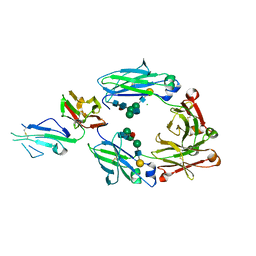 | | Crystal structure of fucosylated IgG1 Fc complexed with bis-glycosylated soluble form of Fc gamma receptor IIIa | | Descriptor: | CHLORIDE ION, Immunoglobulin gamma-1 heavy chain, Low affinity immunoglobulin gamma Fc region receptor III-A, ... | | Authors: | Sakae, Y, Satoh, T, Yagi, H, Yanaka, S, Yamaguchi, T, Isoda, Y, Iida, S, Okamoto, Y, Kato, K. | | Deposit date: | 2017-05-01 | | Release date: | 2017-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational effects of N-glycan core fucosylation of immunoglobulin G Fc region on its interaction with Fc gamma receptor IIIa.
Sci Rep, 7, 2017
|
|
5XJF
 
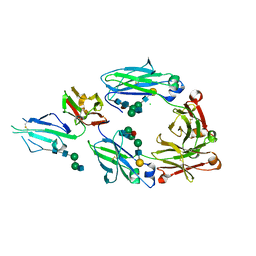 | | Crystal structure of fucosylated IgG Fc Y296W mutant complexed with bis-glycosylated soluble form of Fc gamma receptor IIIa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Sakae, Y, Satoh, T, Yagi, H, Yanaka, S, Yamaguchi, T, Isoda, Y, Iida, S, Okamoto, Y, Kato, K. | | Deposit date: | 2017-05-01 | | Release date: | 2017-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational effects of N-glycan core fucosylation of immunoglobulin G Fc region on its interaction with Fc gamma receptor IIIa.
Sci Rep, 7, 2017
|
|
3AGT
 
 | | Hemerythrin-like domain of DcrH (met) | | Descriptor: | CHLORO DIIRON-OXO MOIETY, Hemerythrin-like domain protein DcrH | | Authors: | Onoda, A, Okamoto, Y, Sugimoto, H, Mizohata, E, Inoue, T, Shiro, Y, Hayashi, T. | | Deposit date: | 2010-04-06 | | Release date: | 2011-04-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Characteristics of Diiron Site with Large Cavity in Hemerythrin-like Domain of DcrH
to be published
|
|
3AGU
 
 | | Hemerythrin-like domain of DcrH (semimet-R) | | Descriptor: | CHLORO DIIRON-OXO MOIETY, Hemerythrin-like domain protein DcrH | | Authors: | Onoda, A, Okamoto, Y, Sugimoto, H, Mizohata, E, Inoue, T, Shiro, Y, Hayashi, T. | | Deposit date: | 2010-04-06 | | Release date: | 2011-04-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Characteristics of Diiron Site with Large Cavity in Hemerythrin-like Domain of DcrH
to be published
|
|
5H5A
 
 | | Mdm12 from K. lactis (1-239), Lys residues are uniformly dimethyl modified | | Descriptor: | Mitochondrial distribution and morphology protein 12, POTASSIUM ION, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Kawano, S, Quinbara, S, Endo, T. | | Deposit date: | 2016-11-04 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure-function insights into direct lipid transfer between membranes by Mmm1-Mdm12 of ERMES
J. Cell Biol., 217, 2018
|
|
5H54
 
 | | Mdm12 from K. lactis 1-239 | | Descriptor: | Mitochondrial distribution and morphology protein 12 | | Authors: | Kawano, S, Quinbara, S, Endo, T. | | Deposit date: | 2016-11-04 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-function insights into direct lipid transfer between membranes by Mmm1-Mdm12 of ERMES
J. Cell Biol., 217, 2018
|
|
5H55
 
 | | Mdm12 from K. lactis | | Descriptor: | Mitochondrial distribution and morphology protein 12 | | Authors: | Kawano, S, Quinbara, S. | | Deposit date: | 2016-11-04 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure-function insights into direct lipid transfer between membranes by Mmm1-Mdm12 of ERMES
J. Cell Biol., 217, 2018
|
|
5H5C
 
 | |
6L9W
 
 | | Crystal structure of mouse TIFA (T9E/C36S mutant) | | Descriptor: | TRAF-interacting protein with FHA domain-containing protein A | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2019-11-11 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural analysis of TIFA: Insight into TIFA-dependent signal transduction in innate immunity.
Sci Rep, 10, 2020
|
|
6L9V
 
 | | Crystal structure of mouse TIFA (T9D/C36S mutant) | | Descriptor: | TRAF-interacting protein with FHA domain-containing protein A | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2019-11-11 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural analysis of TIFA: Insight into TIFA-dependent signal transduction in innate immunity.
Sci Rep, 10, 2020
|
|
6L9U
 
 | | Crystal structure of mouse TIFA | | Descriptor: | TRAF-interacting protein with FHA domain-containing protein A | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2019-11-11 | | Release date: | 2020-04-01 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural analysis of TIFA: Insight into TIFA-dependent signal transduction in innate immunity.
Sci Rep, 10, 2020
|
|
3AGC
 
 | | F218V mutant of the substrate-bound red chlorophyll catabolite reductase from Arabidopsis thaliana | | Descriptor: | 3-{(2Z,3S,4S)-5-[(Z)-(4-ethenyl-3-methyl-5-oxo-1,5-dihydro-2H-pyrrol-2-ylidene)methyl]-2-[(5R)-2-[(3-ethyl-5-formyl-4-methyl-1H-pyrrol-2-yl)methyl]-5-(methoxycarbonyl)-3-methyl-4-oxo-4,5-dihydrocyclopenta[b]pyrrol-6(1H)-ylidene]-4-methyl-3,4-dihydro-2H-pyrrol-3-yl}propanoic acid, Red chlorophyll catabolite reductase, chloroplastic, ... | | Authors: | Sugishima, M, Fukuyama, K. | | Deposit date: | 2010-03-30 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the substrate-bound forms of red chlorophyll catabolite reductase: implications for site-specific and stereospecific reaction
J.Mol.Biol., 402, 2010
|
|
3AGA
 
 | | Crystal structure of RCC-bound red chlorophyll catabolite reductase from Arabidopsis thaliana | | Descriptor: | 3-{(2Z,3S,4S)-5-[(Z)-(4-ethenyl-3-methyl-5-oxo-1,5-dihydro-2H-pyrrol-2-ylidene)methyl]-2-[(5R)-2-[(3-ethyl-5-formyl-4-methyl-1H-pyrrol-2-yl)methyl]-5-(methoxycarbonyl)-3-methyl-4-oxo-4,5-dihydrocyclopenta[b]pyrrol-6(1H)-ylidene]-4-methyl-3,4-dihydro-2H-pyrrol-3-yl}propanoic acid, Red chlorophyll catabolite reductase, chloroplastic, ... | | Authors: | Sugishima, M, Fukuyama, K. | | Deposit date: | 2010-03-30 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of the substrate-bound forms of red chlorophyll catabolite reductase: implications for site-specific and stereospecific reaction
J.Mol.Biol., 402, 2010
|
|
3AGB
 
 | |
6JSA
 
 | |
6JS9
 
 | |
6JSC
 
 | |
6JSB
 
 | |
6JSD
 
 | |
