3ABK
 
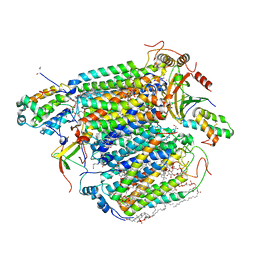 | | Bovine heart cytochrome c oxidase at the NO-bound fully reduced state (50K) | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Ohta, K, Muramoto, K, Shinzawa-Itoh, K, Yamashita, E, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2009-12-16 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of the NO-bound Cu(B) in bovine cytochrome c oxidase
Acta Crystallogr.,Sect.F, 66, 2010
|
|
5GZT
 
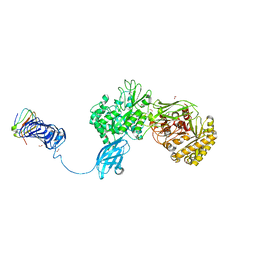 | | Crystal Structure of Chitinase ChiW from Paenibacillus sp. str. FPU-7 Reveals a Novel Type of Bacterial Cell-Surface-Expressed Multi-Modular Enzyme Machinery | | Descriptor: | Chitinase, FORMIC ACID, PHOSPHATE ION, ... | | Authors: | Itoh, T, Hibi, T, Suzuki, F, Sugimoto, I, Fujiwara, A, Inaka, K, Tanaka, H, Ohta, K, Fujii, Y, Taketo, A, Kimoto, H. | | Deposit date: | 2016-10-01 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Chitinase ChiW from Paenibacillus sp. str. FPU-7 Reveals a Novel Type of Bacterial Cell-Surface-Expressed Multi-Modular Enzyme Machinery
PLoS ONE, 11, 2016
|
|
5GZU
 
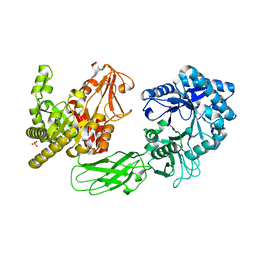 | | Crystal Structure of Chitinase ChiW from Paenibacillus sp. str. FPU-7 Reveals a Novel Type of Bacterial Cell-Surface-Expressed Multi-Modular Enzyme Machinery | | Descriptor: | Chitinase, PHOSPHATE ION | | Authors: | Itoh, T, Hibi, T, Suzuki, F, Sugimoto, I, Fujiwara, A, Inaka, K, Tanaka, H, Ohta, K, Fujii, Y, Taketo, A, Kimoto, H. | | Deposit date: | 2016-10-01 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal Structure of Chitinase ChiW from Paenibacillus sp. str. FPU-7 Reveals a Novel Type of Bacterial Cell-Surface-Expressed Multi-Modular Enzyme Machinery
PLoS ONE, 11, 2016
|
|
5GZV
 
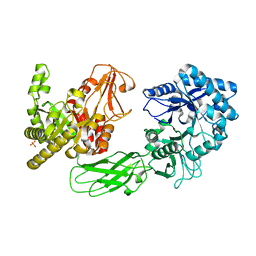 | | Crystal Structure of Chitinase ChiW from Paenibacillus sp. str. FPU-7 Reveals a Novel Type of Bacterial Cell-Surface-Expressed Multi-Modular Enzyme Machinery | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase, PHOSPHATE ION | | Authors: | Itoh, T, Hibi, T, Suzuki, F, Sugimoto, I, Fujiwara, A, Inaka, K, Tanaka, H, Ohta, K, Fujii, Y, Taketo, A, Kimoto, H. | | Deposit date: | 2016-10-01 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal Structure of Chitinase ChiW from Paenibacillus sp. str. FPU-7 Reveals a Novel Type of Bacterial Cell-Surface-Expressed Multi-Modular Enzyme Machinery
PLoS ONE, 11, 2016
|
|
5YYP
 
 | | Structure K137A thaumatin | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Preprothaumatin I | | Authors: | Masuda, T, Kigo, S, Mitsumoto, M, Ohta, K, Suzuki, M, Mikami, B, Kitabatake, N, Tani, F. | | Deposit date: | 2017-12-10 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Positive Charges on the Surface of Thaumatin Are Crucial for the Multi-Point Interaction with the Sweet Receptor.
Front Mol Biosci, 5, 2018
|
|
5YYQ
 
 | | Structure K78A thaumatin | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Preprothaumatin I | | Authors: | Masuda, T, Kigo, S, Mitsumoto, M, Ohta, K, Suzuki, M, Mikami, B, Kitabatake, N, Tani, F. | | Deposit date: | 2017-12-10 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Positive Charges on the Surface of Thaumatin Are Crucial for the Multi-Point Interaction with the Sweet Receptor.
Front Mol Biosci, 5, 2018
|
|
5YYR
 
 | | Structure K106A thaumatin | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Preprothaumatin I | | Authors: | Masuda, T, Kigo, S, Ohta, K, Mitsumoto, M, Mikami, B, Suzuki, M, Kitabatake, N, Tani, F. | | Deposit date: | 2017-12-10 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Positive Charges on the Surface of Thaumatin Are Crucial for the Multi-Point Interaction with the Sweet Receptor.
Front Mol Biosci, 5, 2018
|
|
4Y04
 
 | | Crystal structure of dipeptidyl peptidase 11 (DPP11) from Porphyromonas gingivalis (Space) | | Descriptor: | GLYCEROL, POTASSIUM ION, Peptidase S46 | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Inaka, K, Tanaka, H, Yamada, M, Ohta, K, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2015-02-05 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural and mutational analyses of dipeptidyl peptidase 11 from Porphyromonas gingivalis reveal the molecular basis for strict substrate specificity.
Sci Rep, 5, 2015
|
|
3AG1
 
 | | Bovine Heart Cytochrome c Oxidase in the Carbon Monoxide-bound Fully Reduced State at 280 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Muramoto, K, Ohta, K, Shinzawa-Itoh, K, Kanda, K, Taniguchi, M, Nabekura, H, Yamashita, E, Tsukihara, T, Yoshikawa, S. | | Deposit date: | 2010-03-19 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bovine cytochrome c oxidase structures enable O2 reduction with minimization of reactive oxygens and provide a proton-pumping gate
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3AG4
 
 | | Bovine Heart Cytochrome c Oxidase in the Cyanide Ion-bound Fully Reduced State at 100 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Muramoto, K, Ohta, K, Shinzawa-Itoh, K, Kanda, K, Taniguchi, M, Nabekura, H, Yamashita, E, Tsukihara, T, Yoshikawa, S. | | Deposit date: | 2010-03-19 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Bovine cytochrome c oxidase structures enable O2 reduction with minimization of reactive oxygens and provide a proton-pumping gate
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3AG3
 
 | | Bovine Heart Cytochrome c Oxidase in the Nitric Oxide-bound Fully Reduced State at 100 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Muramoto, K, Ohta, K, Shinzawa-Itoh, K, Kanda, K, Taniguchi, M, Nabekura, H, Yamashita, E, Tsukihara, T, Yoshikawa, S. | | Deposit date: | 2010-03-19 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bovine cytochrome c oxidase structures enable O2 reduction with minimization of reactive oxygens and provide a proton-pumping gate
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3AG2
 
 | | Bovine Heart Cytochrome c Oxidase in the Carbon Monoxide-bound Fully Reduced State at 100 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Muramoto, K, Ohta, K, Shinzawa-Itoh, K, Kanda, K, Taniguchi, M, Nabekura, H, Yamashita, E, Tsukihara, T, Yoshikawa, S. | | Deposit date: | 2010-03-19 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Bovine cytochrome c oxidase structures enable O2 reduction with minimization of reactive oxygens and provide a proton-pumping gate
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2ZJB
 
 | | Crystal structure of the human Dmc1-M200V polymorphic variant | | Descriptor: | Meiotic recombination protein DMC1/LIM15 homolog | | Authors: | Hikiba, J, Hirota, K, Kagawa, W, Ikawa, S, Kinebuchi, T, Sakane, I, Takizawa, Y, Yokoyama, S, Mandon-Pepin, B, Nicolas, A, Shibata, T, Ohta, K, Kurumizaka, H. | | Deposit date: | 2008-03-02 | | Release date: | 2008-08-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural and functional analyses of the DMC1-M200V polymorphism found in the human population
Nucleic Acids Res., 36, 2008
|
|
3X2L
 
 | | X-ray structure of PcCel45A apo form at 95K. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-methylpentane-1,5-diol, Endoglucanase V-like protein | | Authors: | Nakamura, A, Ishida, T, Ohta, K, Tanaka, H, Inaka, K, Samejima, M, Igarashi, K. | | Deposit date: | 2014-12-22 | | Release date: | 2015-10-14 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (0.83 Å) | | Cite: | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
8IJN
 
 | | Bovine Heart Cytochrome c Oxidase in the Nitric Oxide-Bound Fully Reduced State at 100 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Tsukihara, T, Shimada, A, Muramoto, K. | | Deposit date: | 2023-02-27 | | Release date: | 2023-04-12 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bovine cytochrome c oxidase structures enable O2 reduction with minimization of reactive oxygens and provide a proton-pumping gate.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6JTB
 
 | | Crystal structure of dipeptidyl peptidase 11 (DPP11) with citrate from Porphyromonas gingivalis (Space) | | Descriptor: | Asp/Glu-specific dipeptidyl-peptidase, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Roppongi, S, Kushibiki, C, Nakamura, A, Ogasawara, W, Tanaka, N. | | Deposit date: | 2019-04-10 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Fragment-based discovery of the first nonpeptidyl inhibitor of an S46 family peptidase.
Sci Rep, 9, 2019
|
|
6JTC
 
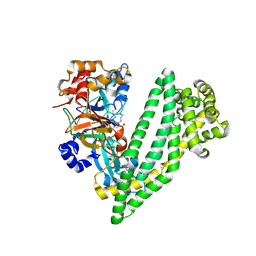 | | Crystal structure of dipeptidyl peptidase 11 (DPP11) with SH-5 from Porphyromonas gingivalis (Space) | | Descriptor: | 2-(2-azanylethylamino)-5-nitro-benzoic acid, Asp/Glu-specific dipeptidyl-peptidase, GLYCEROL | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Roppongi, S, Kushibiki, C, Nakamura, A, Ogasawara, W, Tanaka, N. | | Deposit date: | 2019-04-10 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Fragment-based discovery of the first nonpeptidyl inhibitor of an S46 family peptidase.
Sci Rep, 9, 2019
|
|
4ZM7
 
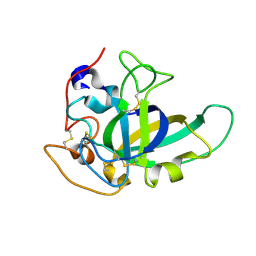 | |
4XZY
 
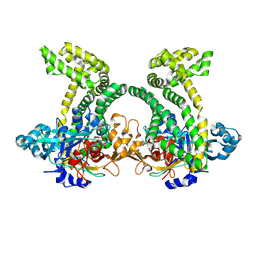 | | Crystal structure of dipeptidyl peptidase 11 (DPP11) from Porphyromonas gingivalis | | Descriptor: | GLYCEROL, Peptidase S46 | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2015-02-05 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and mutational analyses of dipeptidyl peptidase 11 from Porphyromonas gingivalis reveal the molecular basis for strict substrate specificity.
Sci Rep, 5, 2015
|
|
4Y01
 
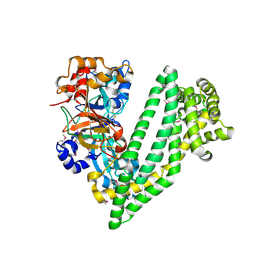 | | Crystal structure of dipeptidyl peptidase 11 (DPP11) from Porphyromonas gingivalis | | Descriptor: | GLYCEROL, Peptidase S46 | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2015-02-05 | | Release date: | 2015-07-15 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural and mutational analyses of dipeptidyl peptidase 11 from Porphyromonas gingivalis reveal the molecular basis for strict substrate specificity.
Sci Rep, 5, 2015
|
|
4Y02
 
 | | Crystal structure of dipeptidyl peptidase 11 (DPP11) from Porphyromonas gingivalis (Ground) | | Descriptor: | GLYCEROL, POTASSIUM ION, Peptidase S46 | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2015-02-05 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural and mutational analyses of dipeptidyl peptidase 11 from Porphyromonas gingivalis reveal the molecular basis for strict substrate specificity.
Sci Rep, 5, 2015
|
|
4Y06
 
 | | Crystal structure of the DAP BII (G675R) dipeptide complex | | Descriptor: | Dipeptidyl aminopeptidase BII, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Sakamoto, Y, Iizuka, I, Tateoka, C, Roppongi, S, Fujimoto, M, Nonaka, T, Ogasawara, W, Tanaka, N. | | Deposit date: | 2015-02-05 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural and mutational analyses of dipeptidyl peptidase 11 from Porphyromonas gingivalis reveal the molecular basis for strict substrate specificity.
Sci Rep, 5, 2015
|
|
3X2M
 
 | | X-ray structure of PcCel45A with cellopentaose at 0.64 angstrom resolution. | | Descriptor: | Endoglucanase V-like protein, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Nakamura, A, Ishida, T, Samejima, M, Igarashi, K. | | Deposit date: | 2014-12-22 | | Release date: | 2015-10-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (0.64 Å) | | Cite: | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
3X2P
 
 | | Neutron and X-ray joint refined structure of PcCel45A with cellopentaose at 298K. | | Descriptor: | Endoglucanase V-like protein, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Nakamura, A, Ishida, T, Kusaka, K, Yamada, T, Tanaka, I, Niimura, N, Samejima, M, Igarashi, K. | | Deposit date: | 2014-12-22 | | Release date: | 2015-10-14 | | Last modified: | 2020-07-29 | | Method: | NEUTRON DIFFRACTION (1.518 Å), X-RAY DIFFRACTION | | Cite: | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
3X2O
 
 | | Neutron and X-ray joint refined structure of PcCel45A apo form at 298K. | | Descriptor: | Endoglucanase V-like protein | | Authors: | Nakamura, A, Ishida, T, Kusaka, K, Yamada, T, Tanaka, I, Niimura, N, Samejima, M, Igarashi, K. | | Deposit date: | 2014-12-22 | | Release date: | 2015-10-07 | | Last modified: | 2019-12-18 | | Method: | NEUTRON DIFFRACTION (1.5 Å), X-RAY DIFFRACTION | | Cite: | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
