2V6X
 
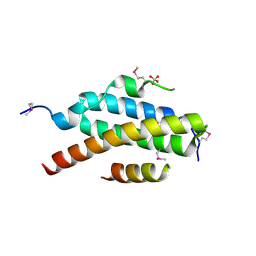 | | Stractural insight into the interaction between ESCRT-III and Vps4 | | Descriptor: | DOA4-INDEPENDENT DEGRADATION PROTEIN 4, SULFATE ION, VACUOLAR PROTEIN SORTING-ASSOCIATED PROTEIN 4 | | Authors: | Obita, T, Perisic, O, Ghazi-Tabatabai, S, Saksena, S, Emr, S.D, Williams, R.L. | | Deposit date: | 2007-07-23 | | Release date: | 2007-10-16 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Basis for Selective Recognition of Escrt-III by the Aaa ATPase Vps4
Nature, 449, 2007
|
|
2V6Y
 
 | | Structure of the MIT domain from a S. solfataricus Vps4-like ATPase | | Descriptor: | AAA FAMILY ATPASE, P60 KATANIN, S,R MESO-TARTARIC ACID | | Authors: | Obita, T, Saksena, S, Ghazi-Tabatabai, S, Gill, D.J, Perisic, O, Emr, S.D, Williams, R.L. | | Deposit date: | 2007-07-24 | | Release date: | 2007-10-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Selective Recognition of Escrt-III by the Aaa ATPase Vps4
Nature, 449, 2007
|
|
1IVM
 
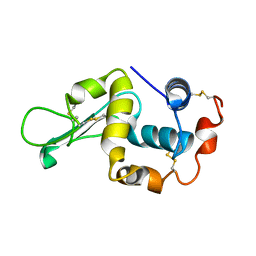 | |
5X2L
 
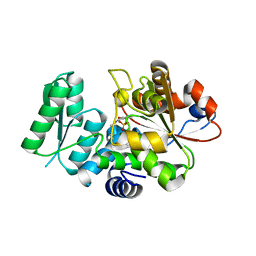 | | Crystal Structure of Human Serine Racemase | | Descriptor: | MAGNESIUM ION, PYRIDOXAL-5'-PHOSPHATE, Serine racemase | | Authors: | Obita, T, Matsumoto, K, Mori, H, Toyooka, N, Mizuguchi, M. | | Deposit date: | 2017-02-01 | | Release date: | 2018-01-31 | | Last modified: | 2021-02-24 | | Method: | X-RAY DIFFRACTION (1.806 Å) | | Cite: | Design, synthesis, and evaluation of novel inhibitors for wild-type human serine racemase.
Bioorg. Med. Chem. Lett., 2017
|
|
5AON
 
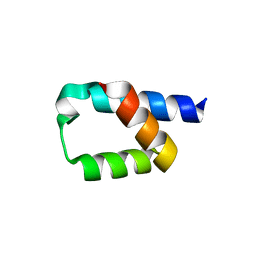 | | Crystal structure of the conserved N-terminal domain of Pex14 from Trypanosoma brucei | | Descriptor: | PEROXIN 14, SULFATE ION | | Authors: | Obita, T, Sugawara, Y, Mizuguchi, M, Watanabe, Y, Kawaguchi, K, Imanaka, T. | | Deposit date: | 2015-09-11 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.646 Å) | | Cite: | Characterization of the Interaction between Trypanosoma Brucei Pex5P and its Receptor Pex14P.
FEBS Lett., 590, 2016
|
|
7X4O
 
 | |
2V1T
 
 | | CRYSTAL STRUCTURE OF RAT TOM20-ALDH PRESEQUENCE COMPLEX | | Descriptor: | ALDEHYDE DEHYDROGENASE, MITOCHONDRIAL IMPORT RECEPTOR SUBUNIT TOM20 HOMOLOG | | Authors: | Obita, T, Igura, M, Ose, T, Endo, T, Maenaka, K, Kohda, D. | | Deposit date: | 2007-05-29 | | Release date: | 2007-06-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Tom20 Recognizes Mitochondrial Presequences Through Dynamic Equilibrium Among Multiple Bound States.
Embo J., 26, 2007
|
|
2V1S
 
 | | CRYSTAL STRUCTURE OF RAT TOM20-ALDH PRESEQUENCE COMPLEX | | Descriptor: | ALDEHYDE DEHYDROGENASE, MITOCHONDRIAL IMPORT RECEPTOR SUBUNIT TOM20 HOMOLOG | | Authors: | Obita, T, Igura, M, Ose, T, Endo, T, Maenaka, K, Kohda, D. | | Deposit date: | 2007-05-29 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Tom20 Recognizes Mitochondrial Presequences Through Dynamic Equilibrium Among Multiple Bound States.
Embo J., 26, 2007
|
|
5GPY
 
 | | Crystal structure of the human TFIIE complex | | Descriptor: | General transcription factor IIE subunit 1, Transcription initiation factor IIE subunit beta, ZINC ION | | Authors: | Obita, T, Miwa, K, Kojima, R, Ohkuma, Y, Tamura, Y, Mizuguchi, M. | | Deposit date: | 2016-08-05 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Human General Transcription Factor TFIIE at Atomic Resolution
J.Mol.Biol., 428, 2016
|
|
2W2U
 
 | | STRUCTURAL INSIGHT INTO THE INTERACTION BETWEEN ARCHAEAL ESCRT-III AND AAA-ATPASE | | Descriptor: | CONSERVED ARCHAEAL PROTEIN, HYPOTHETICAL P60 KATANIN | | Authors: | Obita, T, Samson, R.Y, Perisic, O, Freund, S.M, Bell, S.D, Williams, R.L. | | Deposit date: | 2008-11-04 | | Release date: | 2009-07-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Role for the Escrt System in Cell Division in Archaea.
Science, 322, 2008
|
|
8I75
 
 | |
8I76
 
 | |
8I74
 
 | |
8JNU
 
 | |
8JOK
 
 | |
8JQW
 
 | |
4BWS
 
 | | Crystal structure of the heterotrimer of PQBP1, U5-15kD and U5-52kD. | | Descriptor: | CD2 ANTIGEN CYTOPLASMIC TAIL-BINDING PROTEIN 2, POLYGLUTAMINE-BINDING PROTEIN 1, THIOREDOXIN-LIKE PROTEIN 4A | | Authors: | Mizuguchi, M, Obita, T, Serita, T, Kojima, R, Morimoto, T, Nabeshima, Y, Okazawa, H. | | Deposit date: | 2013-07-04 | | Release date: | 2014-04-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutations in the Pqbp1 Gene Prevent its Interaction with the Spliceosomal Protein U5-15Kd.
Nat.Commun., 5, 2014
|
|
4BWQ
 
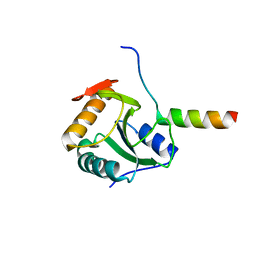 | | Crystal structure of U5-15kD in a complex with PQBP1 | | Descriptor: | POLYGLUTAMINE-BINDING PROTEIN 1, THIOREDOXIN-LIKE PROTEIN 4A | | Authors: | Mizuguchi, M, Obita, T, Serita, T, Kojima, R, Morimoto, T, Nabeshima, Y, Okazawa, H. | | Deposit date: | 2013-07-04 | | Release date: | 2014-04-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mutations in the Pqbp1 Gene Prevent its Interaction with the Spliceosomal Protein U5-15Kd.
Nat.Commun., 5, 2014
|
|
4A5Z
 
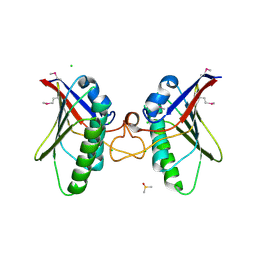 | | Structures of MITD1 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, MIT DOMAIN-CONTAINING PROTEIN 1 | | Authors: | Hadders, M.A, Agromayor, M, Caballe, A, Obita, T, Perisic, O, Williams, R.L, Martin-Serrano, J. | | Deposit date: | 2011-10-30 | | Release date: | 2012-11-14 | | Last modified: | 2013-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Escrt-III Binding Protein Mitd1 is Involved in Cytokinesis and Has an Unanticipated Pld Fold that Binds Membranes.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4CDO
 
 | | Crystal structure of PQBP1 bound to spliceosomal U5-15kD | | Descriptor: | THIOREDOXIN-LIKE PROTEIN 4A, POLYGLUTAMINE-BINDING PROTEIN | | Authors: | Mizuguchi, M, Obita, T, Serita, T, Kojima, R, Nabeshima, Y, Okazawa, H. | | Deposit date: | 2013-11-05 | | Release date: | 2014-04-30 | | Last modified: | 2014-05-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutations in the Pqbp1 Gene Prevent its Interaction with the Spliceosomal Protein U5-15Kd.
Nat.Commun., 5, 2014
|
|
7WL6
 
 | |
5H0Z
 
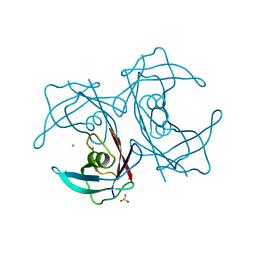 | | Crystal structure of P113A mutated human transthyretin | | Descriptor: | CHLORIDE ION, SULFATE ION, Transthyretin | | Authors: | Yokoyama, T, Hanawa, Y, Obita, T, Mizuguchi, M. | | Deposit date: | 2016-10-07 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.735 Å) | | Cite: | Stability and crystal structures of His88 mutant human transthyretins
FEBS Lett., 591, 2017
|
|
5H0X
 
 | |
5H0V
 
 | | Crystal structure of H88A mutated human transthyretin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, MAGNESIUM ION, Transthyretin | | Authors: | Yokoyama, T, Hanawa, Y, Obita, T, Mizuguchi, M. | | Deposit date: | 2016-10-07 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Stability and crystal structures of His88 mutant human transthyretins
FEBS Lett., 591, 2017
|
|
5H0W
 
 | | Crystal structure of H88F mutated human transthyretin | | Descriptor: | SULFATE ION, Transthyretin | | Authors: | Yokoyama, T, Hanawa, Y, Obita, T, Mizuguchi, M. | | Deposit date: | 2016-10-07 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Stability and crystal structures of His88 mutant human transthyretins
FEBS Lett., 591, 2017
|
|
