1A5E
 
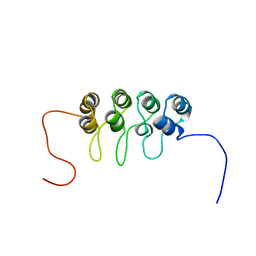 | | SOLUTION NMR STRUCTURE OF TUMOR SUPPRESSOR P16INK4A, 18 STRUCTURES | | Descriptor: | TUMOR SUPPRESSOR P16INK4A | | Authors: | Byeon, I.-J.L, Li, J, Ericson, K, Selby, T.L, Tevelev, A, Kim, H.-J, O'Maille, P, Tsai, M.-D. | | Deposit date: | 1998-02-13 | | Release date: | 1999-08-13 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Tumor suppressor p16INK4A: determination of solution structure and analyses of its interaction with cyclin-dependent kinase 4.
Mol.Cell, 1, 1998
|
|
2A5E
 
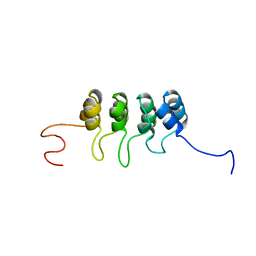 | | SOLUTION NMR STRUCTURE OF TUMOR SUPPRESSOR P16INK4A, RESTRAINED MINIMIZED MEAN STRUCTURE | | Descriptor: | TUMOR SUPPRESSOR P16INK4A | | Authors: | Byeon, I.-J.L, Li, J, Ericson, K, Selby, T.L, Tevelev, A, Kim, H.-J, O'Maille, P, Tsai, M.-D. | | Deposit date: | 1998-02-13 | | Release date: | 1999-08-13 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Tumor suppressor p16INK4A: determination of solution structure and analyses of its interaction with cyclin-dependent kinase 4.
Mol.Cell, 1, 1998
|
|
1BU9
 
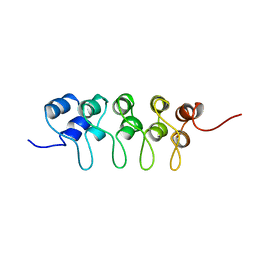 | | SOLUTION STRUCTURE OF P18-INK4C, 21 STRUCTURES | | Descriptor: | PROTEIN (CYCLIN-DEPENDENT KINASE 6 INHIBITOR) | | Authors: | Byeon, I.-J.L, Li, J, Tsai, M.-D. | | Deposit date: | 1998-09-15 | | Release date: | 1999-09-13 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Tumor suppressor INK4: determination of the solution structure of p18INK4C and demonstration of the functional significance of loops in p18INK4C and p16INK4A.
Biochemistry, 38, 1999
|
|
8T2M
 
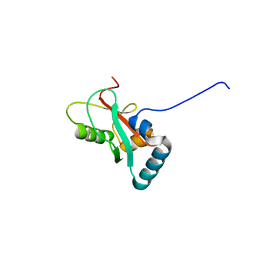 | | Crystal structure of GABARAP in complex with the LIR of NSs4 | | Descriptor: | Non-structural protein S,Gamma-aminobutyric acid receptor-associated protein chimera | | Authors: | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | Deposit date: | 2023-06-06 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | An LIR motif in the Rift Valley fever virus NSs protein is critical for the interaction with LC3 family members and inhibition of autophagy.
Plos Pathog., 20, 2024
|
|
8T2N
 
 | | Crystal structure of GABARAP in complex with the LIR of NSs3 | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein, Non-structural protein S | | Authors: | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | Deposit date: | 2023-06-06 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | An LIR motif in the Rift Valley fever virus NSs protein is critical for the interaction with LC3 family members and inhibition of autophagy.
Plos Pathog., 20, 2024
|
|
8T2L
 
 | | Crystal structure of LC3A in complex with the LIR of NSs4 | | Descriptor: | Non-structural protein S,Microtubule-associated proteins 1A/1B light chain 3A chimera | | Authors: | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | Deposit date: | 2023-06-06 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | An LIR motif in the Rift Valley fever virus NSs protein is critical for the interaction with LC3 family members and inhibition of autophagy.
Plos Pathog., 20, 2024
|
|
1DC2
 
 | | SOLUTION NMR STRUCTURE OF TUMOR SUPPRESSOR P16INK4A, 20 STRUCTURES | | Descriptor: | CYCLIN-DEPENDENT KINASE 4 INHIBITOR A (P16INK4A) | | Authors: | Byeon, I.-J.L, Li, J, Yuan, C, Tsai, M.-D. | | Deposit date: | 1999-11-04 | | Release date: | 1999-12-23 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Tumor suppressor INK4: refinement of p16INK4A structure and determination of p15INK4B structure by comparative modeling and NMR data.
Protein Sci., 9, 2000
|
|
