8FE5
 
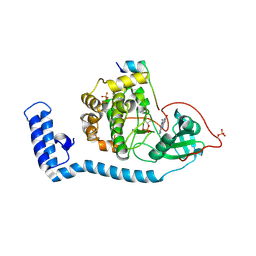 | | Structure of J-PKAc chimera complexed with Aplithianine B | | Descriptor: | 6-[(6P)-6-(1-methyl-1H-imidazol-5-yl)-2,3-dihydro-4H-1,4-thiazin-4-yl]-7,9-dihydro-8H-purin-8-one, DnaJ homolog subfamily B member 1,cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Du, L, Wilson, B.A.P, Li, N, Martinez Fiesco, J.A, Dalilian, M, Wang, D, Smith, E.A, Wamiru, A, Goncharova, E.I, Zhang, P, O'Keefe, B.R. | | Deposit date: | 2022-12-05 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery and Synthesis of a Naturally Derived Protein Kinase Inhibitor that Selectively Inhibits Distinct Classes of Serine/Threonine Kinases.
J.Nat.Prod., 86, 2023
|
|
6XN9
 
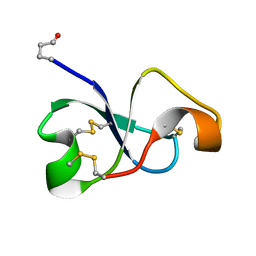 | | Solution NMR structure of recifin, a cysteine-rich tyrosyl-DNA Phosphodiesterase I modulatory peptide from the marine sponge Axinella sp. | | Descriptor: | Recifin modulatory peptide | | Authors: | Schroeder, C.I, Rosengren, K.J, O'Keefe, B.R. | | Deposit date: | 2020-07-02 | | Release date: | 2021-02-10 | | Method: | SOLUTION NMR | | Cite: | Recifin A, Initial Example of the Tyr-Lock Peptide Structural Family, Is a Selective Allosteric Inhibitor of Tyrosyl-DNA Phosphodiesterase I.
J.Am.Chem.Soc., 142, 2020
|
|
8FE2
 
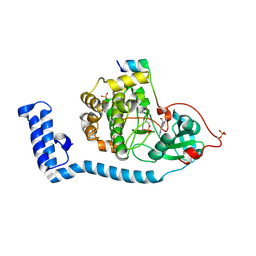 | | Structure of J-PKAc chimera complexed with Aplithianine A | | Descriptor: | 6-[(6M)-6-(1-methyl-1H-imidazol-5-yl)-2,3-dihydro-4H-1,4-thiazin-4-yl]-9H-purine, DnaJ homolog subfamily B member 1, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Du, L, Wilson, B.A.P, Li, N, Dalilian, M, Wang, D, Martinez Fiesco, J.A, Smith, E.A, Wamiru, A, Goncharova, E.I, Zhang, P, O'Keefe, B.R. | | Deposit date: | 2022-12-05 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structures of J-PKAc chimera complexed with Aplithianine and derivatives
To Be Published
|
|
8FEC
 
 | | Structure of J-PKAc chimera complexed with Aplithianine derivative | | Descriptor: | 6-[(6P)-6-(4-bromo-1-methyl-1H-imidazol-5-yl)-2,3-dihydro-4H-1,4-thiazin-4-yl]-7H-purine, DnaJ homolog subfamily B member 1,cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Du, L, Wilson, B.A.P, Li, N, Martinez Fiesco, J.A, Dalilian, M, Wang, D, Smith, E.A, Wamiru, A, Goncharova, E.I, Zhang, P, O'Keefe, B.R. | | Deposit date: | 2022-12-06 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of J-PKAc chimera complexed with Aplithianine and derivatives
To Be Published
|
|
1L5B
 
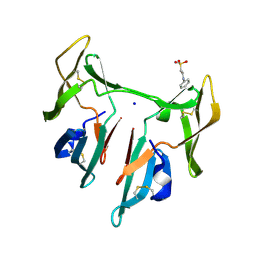 | | DOMAIN-SWAPPED CYANOVIRIN-N DIMER | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, SODIUM ION, cyanovirin-N | | Authors: | Barrientos, L.G, Louis, J.M, Botos, I, Mori, T, Han, Z, O'Keefe, B.R, Boyd, M.R, Wlodawer, A, Gronenborn, A.M. | | Deposit date: | 2002-03-06 | | Release date: | 2002-05-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The domain-swapped dimer of cyanovirin-N is in a metastable folded state: reconciliation of X-ray and NMR structures.
Structure, 10, 2002
|
|
1L5E
 
 | | The domain-swapped dimer of CV-N in solution | | Descriptor: | Cyanovirin-N | | Authors: | Barrientos, L.G, Louis, J.M, Botos, I, Mori, T, Han, Z, O'Keefe, B.R, Boyd, M.R, Wlodawer, A, Gronenborn, A.M. | | Deposit date: | 2002-03-06 | | Release date: | 2002-06-05 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The domain-swapped dimer of cyanovirin-N is in a metastable folded state: reconciliation of X-ray and NMR structures.
Structure, 10, 2002
|
|
3GXZ
 
 | | Crystal structure of cyanovirin-n complexed to oligomannose-9 (man-9) | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Cyanovirin-N, MAGNESIUM ION, ... | | Authors: | Botos, I, O'Keefe, B.R, Shenoy, S.R, Seeberger, P.H, Boyd, M.R, Wlodawer, A. | | Deposit date: | 2009-04-03 | | Release date: | 2009-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the complexes of a potent anti-HIV protein cyanovirin-N and high mannose oligosaccharides
J.Biol.Chem., 277, 2002
|
|
3GXY
 
 | | Crystal structure of cyanovirin-n complexed to a synthetic hexamannoside | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Cyanovirin-N, MAGNESIUM ION, ... | | Authors: | Botos, I, O'Keefe, B.R, Shenoy, S.R, Seeberger, P.H, Boyd, M.R, Wlodawer, A. | | Deposit date: | 2009-04-03 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of the complexes of a potent anti-HIV protein cyanovirin-n and high mannose oligosaccharides
J.Biol.Chem., 277, 2002
|
|
5DUY
 
 | | Structure of lectin from the sea mussel Crenomytilus grayanus | | Descriptor: | GLYCEROL, GalNAc/Gal-specific lectin | | Authors: | Lubkowski, J, Jakob, M, O'Keefe, B, Wlodawer, A. | | Deposit date: | 2015-09-21 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure of a lectin from the sea mussel Crenomytilus grayanus (CGL).
Acta Crystallogr.,Sect.F, 71, 2015
|
|
2NUO
 
 | |
2NU5
 
 | |
2QSK
 
 | | Atomic-resolution crystal structure of the Recombinant form of Scytovirin | | Descriptor: | CHLORIDE ION, GLYCEROL, scytovirin | | Authors: | Moulaei, T, Botos, I, Ziolkowska, N.E, Dauter, Z, Wlodawer, A. | | Deposit date: | 2007-07-31 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic-resolution crystal structure of the antiviral lectin scytovirin.
Protein Sci., 16, 2007
|
|
2QT4
 
 | | Atomic-resolution crystal structure of the natural form of Scytovirin | | Descriptor: | scytovirin | | Authors: | Moulaei, T, Botos, I, Ziolkowska, N.E, Dauter, Z, Wlodawer, A. | | Deposit date: | 2007-08-01 | | Release date: | 2007-11-27 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Atomic-resolution crystal structure of the antiviral lectin scytovirin.
Protein Sci., 16, 2007
|
|
2GTY
 
 | | Crystal structure of unliganded griffithsin | | Descriptor: | 1,2-ETHANEDIOL, Griffithsin, SULFATE ION | | Authors: | Ziolkowska, N.E, Wlodawer, A. | | Deposit date: | 2006-04-28 | | Release date: | 2006-08-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Domain-swapped structure of the potent antiviral protein griffithsin and its mode of carbohydrate binding.
Structure, 14, 2006
|
|
2GUE
 
 | |
2HYR
 
 | | Crystal structure of a complex of griffithsin with maltose | | Descriptor: | 1,2-ETHANEDIOL, Griffithsin, MAGNESIUM ION, ... | | Authors: | Ziolkowska, N.E, Wlodawer, A. | | Deposit date: | 2006-08-07 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystallographic, thermodynamic, and molecular modeling studies of the mode of binding of oligosaccharides to the potent antiviral protein griffithsin.
Proteins, 67, 2007
|
|
3LKY
 
 | | Monomeric Griffithsin with a Single Gly-Ser Insertion | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Griffithsin, ... | | Authors: | Moulaei, T, Wlodawer, A. | | Deposit date: | 2010-01-28 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Monomerization of viral entry inhibitor griffithsin elucidates the relationship between multivalent binding to carbohydrates and anti-HIV activity.
Structure, 18, 2010
|
|
3LL2
 
 | |
3LL0
 
 | | Monomeric Griffithsin with two Gly-Ser Insertions | | Descriptor: | GLYCEROL, Griffithsin, SULFATE ION | | Authors: | Moulaei, T, Wlodawer, A. | | Deposit date: | 2010-01-28 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Monomerization of viral entry inhibitor griffithsin elucidates the relationship between multivalent binding to carbohydrates and anti-HIV activity.
Structure, 18, 2010
|
|
3LL1
 
 | |
2GUC
 
 | |
2GUD
 
 | |
2HYQ
 
 | |
2GUX
 
 | | Selenomethionine derivative of griffithsin | | Descriptor: | SULFATE ION, griffithsin | | Authors: | Ziolkowska, N.E, Wlodawer, A. | | Deposit date: | 2006-05-01 | | Release date: | 2006-08-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Domain-swapped structure of the potent antiviral protein griffithsin and its mode of carbohydrate binding.
Structure, 14, 2006
|
|
2JMV
 
 | |
