4HQJ
 
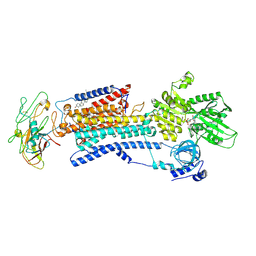 | | Crystal structure of Na+,K+-ATPase in the Na+-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHOLESTEROL, MAGNESIUM ION, ... | | Authors: | Nyblom, M, Reinhard, L, Gourdon, P, Nissen, P. | | Deposit date: | 2012-10-25 | | Release date: | 2013-10-02 | | Last modified: | 2014-09-10 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Crystal structure of Na+, K(+)-ATPase in the Na(+)-bound state.
Science, 342, 2013
|
|
3CLL
 
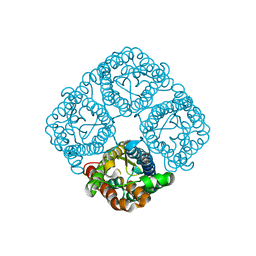 | | Crystal structure of the Spinach Aquaporin SoPIP2;1 S115E mutant | | Descriptor: | Aquaporin | | Authors: | Nyblom, M, Alfredsson, A, Hallgren, K, Hedfalk, K, Neutze, R, Trnroth-Horsefield, S. | | Deposit date: | 2008-03-19 | | Release date: | 2009-02-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional analysis of SoPIP2;1 mutants adds insight into plant aquaporin gating.
J.Mol.Biol., 387, 2009
|
|
3CN6
 
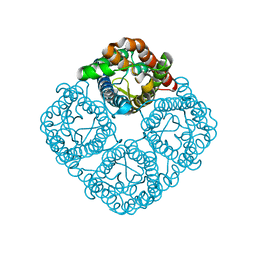 | | Crystal structure of the Spinach Aquaporin SoPIP2;1 S274E mutant | | Descriptor: | Aquaporin, CADMIUM ION | | Authors: | Nyblom, M, Alfredsson, A, Hallgren, K, Hedfalk, K, Neutze, R, Tornroth-Horsefield, S. | | Deposit date: | 2008-03-25 | | Release date: | 2009-02-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural and functional analysis of SoPIP2;1 mutants adds insight into plant aquaporin gating.
J.Mol.Biol., 387, 2009
|
|
3CN5
 
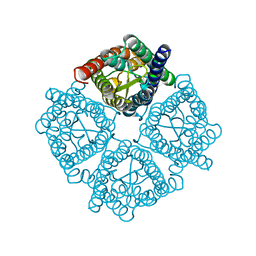 | | Crystal structure of the Spinach Aquaporin SoPIP2;1 S115E, S274E mutant | | Descriptor: | Aquaporin | | Authors: | Nyblom, M, Alfredsson, A, Hallgren, K, Hedfalk, K, Neutze, R, Tornroth-Horsefield, S. | | Deposit date: | 2008-03-25 | | Release date: | 2009-02-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and functional analysis of SoPIP2;1 mutants adds insight into plant aquaporin gating.
J.Mol.Biol., 387, 2009
|
|
7ZOS
 
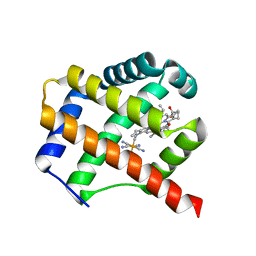 | | Class 1 Phytoglobin from Sugar beet (BvPgb1.2) | | Descriptor: | CYANIDE ION, HEXACYANOFERRATE(3-), Non-symbiotic hemoglobin class 1, ... | | Authors: | Nyblom, M, Christensen, S, Eriksson, N, Bulow, L. | | Deposit date: | 2022-04-26 | | Release date: | 2022-09-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Oxidative Implications of Substituting a Conserved Cysteine Residue in Sugar Beet Phytoglobin BvPgb 1.2.
Antioxidants, 11, 2022
|
|
7Z1U
 
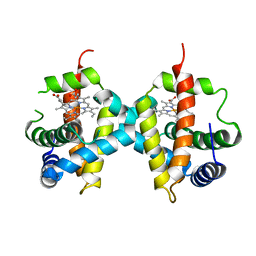 | | Biochemical implications of the substitution of a unique cysteine residue in sugar beet phytoglobin BvPgb 1.2 | | Descriptor: | Non-symbiotic hemoglobin class 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nyblom, M, Christensen, S, Leiva Eriksson, N, Bulow, L. | | Deposit date: | 2022-02-25 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Oxidative Implications of Substituting a Conserved Cysteine Residue in Sugar Beet Phytoglobin BvPgb 1.2.
Antioxidants, 11, 2022
|
|
7QU4
 
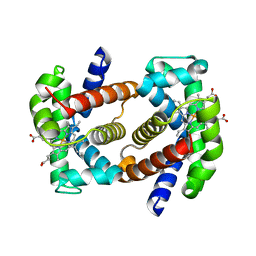 | | Recombinant Human Fetal Hemoglobin mutant - alpha subunit mutations K11E,K56E,N78D,K90E | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit gamma-2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kettisen, K, Nyblom, M, Bulow, L. | | Deposit date: | 2022-01-17 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural and oxidative investigation of a recombinant high-yielding fetal hemoglobin mutant.
Front Mol Biosci, 10, 2023
|
|
4JC6
 
 | | Mercury activation of the plant aquaporin SoPIP2;1 - structural and functional characterization | | Descriptor: | Aquaporin, CADMIUM ION, MERCURY (II) ION, ... | | Authors: | Frick, A, Jarva, M, Nyblom, M, Ekvall, M, Uzdavinys, P, Tornroth-Horsefield, S. | | Deposit date: | 2013-02-21 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Mercury increases water permeability of a plant aquaporin through a non-cysteine-related mechanism.
Biochem.J., 454, 2013
|
|
6ZCT
 
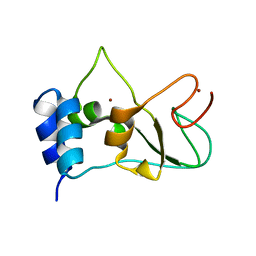 | | Nonstructural protein 10 (nsp10) from SARS CoV-2 | | Descriptor: | ZINC ION, nsp10 | | Authors: | Rogstam, A, Nyblom, M, Christensen, S, Sele, C, Lindvall, T, Rasmussen, A.A, Andre, I, Fisher, S.Z, Knecht, W, Kozielski, F. | | Deposit date: | 2020-06-12 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of Non-Structural Protein 10 from Severe Acute Respiratory Syndrome Coronavirus-2.
Int J Mol Sci, 21, 2020
|
|
7ORR
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00022 | | Descriptor: | 4-PHENYL-1H-IMIDAZOLE, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | Deposit date: | 2021-06-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
7ORV
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00239 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | Deposit date: | 2021-06-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
7ORU
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00221 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | Deposit date: | 2021-06-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
7ORW
 
 | | Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00265 | | Descriptor: | 1H-benzimidazol-4-amine, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Talibov, V.O, Kozielski, F, Sele, C, Lou, J, Dong, D, Wang, Q, Shi, X, Nyblom, M, Rogstam, A, Krojer, T, Knecht, W, Fisher, S.Z. | | Deposit date: | 2021-06-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16.
Rsc Chem Biol, 3, 2022
|
|
6ZPE
 
 | | Nonstructural protein 10 (nsp10) from SARS CoV-2 | | Descriptor: | CHLORIDE ION, GLYCEROL, Replicase polyprotein 1ab, ... | | Authors: | Fisher, S.Z, Kozielski, F. | | Deposit date: | 2020-07-08 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of Non-Structural Protein 10 from Severe Acute Respiratory Syndrome Coronavirus-2.
Int J Mol Sci, 21, 2020
|
|
6RQW
 
 | |
6RQN
 
 | |
6RQU
 
 | |
6RQQ
 
 | |
