7AXZ
 
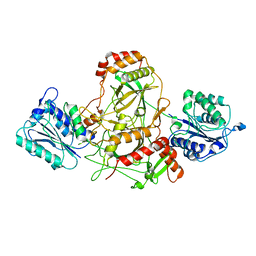 | | Ku70/80 complex apo form | | Descriptor: | X-ray repair cross-complementing protein 5, X-ray repair cross-complementing protein 6 | | Authors: | Hnizda, A, Tesina, P, Novak, P, Blundell, T.L. | | Deposit date: | 2020-11-10 | | Release date: | 2021-02-10 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | SAP domain forms a flexible part of DNA aperture in Ku70/80.
Febs J., 288, 2021
|
|
2G98
 
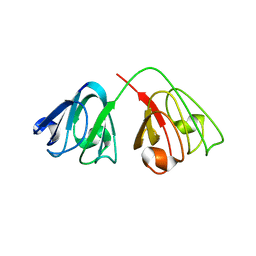 | | human gamma-D-crystallin | | Descriptor: | Gamma crystallin D | | Authors: | Kmoch, S, Brynda, J, Awsav, B, Bezouska, K, Novak, P, Rezacova, P. | | Deposit date: | 2006-03-06 | | Release date: | 2006-05-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Link between a novel human gamma-D-crystallin allele and a unique cataract phenotype explained by protein crystallography.
Hum.Mol.Genet., 12, 2000
|
|
2NAN
 
 | |
2JNT
 
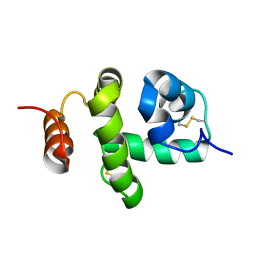 | | Structure of Bombyx mori Chemosensory Protein 1 in Solution | | Descriptor: | Chemosensory protein CSP1 | | Authors: | Jansen, S, Zidek, L, Chmelik, J, Novak, P, Padrta, P, Picimbon, J, Lofstedt, C, Sklenar, V. | | Deposit date: | 2007-02-02 | | Release date: | 2007-11-20 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structure of Bombyx mori chemosensory protein 1 in solution
Arch.Insect Biochem.Physiol., 66, 2007
|
|
2M94
 
 | |
2MTI
 
 | | NMR structure of the lymphocyte receptor NKR-P1A | | Descriptor: | Killer cell lectin-like receptor subfamily B member 1A | | Authors: | Chmelik, J, Rozbesky, D, Pospisilova, E, Adamek, D, Novak, P. | | Deposit date: | 2014-08-19 | | Release date: | 2015-11-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the lymphocyte receptor Nkrp1a reveals a distinct conformation of the long loop region as compared to in the crystal structure.
Proteins, 84, 2016
|
|
8R6T
 
 | | NMR solution structure of thyropin IrThy-Cd from the hard tick Ixodes ricinus | | Descriptor: | Putative two thyropin protein (Fragment) | | Authors: | Srb, P, Veverka, V, Matouskova, Z, Orsaghova, K, Mares, M. | | Deposit date: | 2023-11-23 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-06 | | Method: | SOLUTION NMR | | Cite: | An Unusual Two-Domain Thyropin from Tick Saliva: NMR Solution Structure and Highly Selective Inhibition of Cysteine Cathepsins Modulated by Glycosaminoglycans.
Int J Mol Sci, 25, 2024
|
|
5YI9
 
 | | Solution structure of the LEDGF/p75 IBD - JPO2 (aa 56-91) complex | | Descriptor: | PC4 and SFRS1-interacting protein,Cell division cycle-associated 7-like protein | | Authors: | Veverka, V. | | Deposit date: | 2017-10-03 | | Release date: | 2018-08-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Affinity switching of the LEDGF/p75 IBD interactome is governed by kinase-dependent phosphorylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6TVM
 
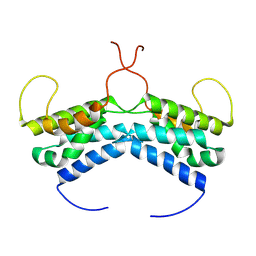 | | LEDGF/p75 dimer (residues 345-467) | | Descriptor: | PC4 and SFRS1-interacting protein | | Authors: | Lux, V, Veverka, V. | | Deposit date: | 2020-01-10 | | Release date: | 2020-09-09 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Molecular Mechanism of LEDGF/p75 Dimerization.
Structure, 28, 2020
|
|
5EDF
 
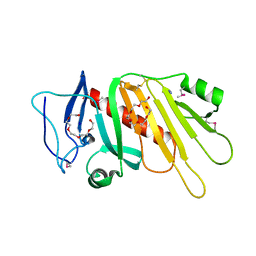 | | Crystal structure of the selenomethionine-substituted iron-regulated protein FrpD from Neisseria meningitidis | | Descriptor: | AZIDE ION, FrpC operon protein, HEXAETHYLENE GLYCOL, ... | | Authors: | Sviridova, E, Bumba, L, Rezacova, P, Sebo, P, Kuta Smatanova, I. | | Deposit date: | 2015-10-21 | | Release date: | 2017-02-01 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of the interaction between the putative adhesion-involved and iron-regulated FrpD and FrpC proteins of Neisseria meningitidis.
Sci Rep, 7, 2017
|
|
5EDJ
 
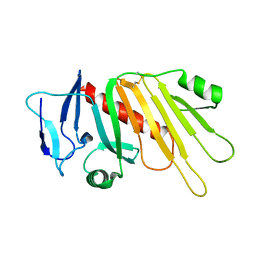 | | Crystal structure of the Neisseria meningitidis iron-regulated outer membrane lipoprotein FrpD | | Descriptor: | FrpC operon protein | | Authors: | Sviridova, E, Bumba, L, Rezacova, P, Sebo, P, Kuta Smatanova, I. | | Deposit date: | 2015-10-21 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of the interaction between the putative adhesion-involved and iron-regulated FrpD and FrpC proteins of Neisseria meningitidis.
Sci Rep, 7, 2017
|
|
8CBQ
 
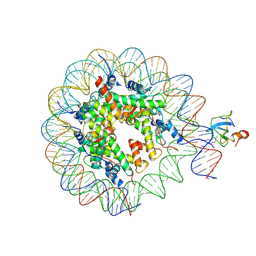 | | structure of LEDGF/p75 PWWP domain bound to the H3K36 trimethylated dinucleosome | | Descriptor: | Histone H2A, Histone H2B 1.1, Histone H3, ... | | Authors: | Koutna, E, Kouba, T, Novacek, J, Veverka, V. | | Deposit date: | 2023-01-25 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Multivalency of nucleosome recognition by LEDGF.
Nucleic Acids Res., 51, 2023
|
|
8CBN
 
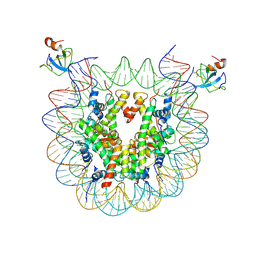 | | structure of LEDGF/p75 PWWP domain bound to the H3K36 trimethylated dinucleosome | | Descriptor: | Histone H2A, Histone H2B 1.1, Histone H3, ... | | Authors: | Koutna, E, Kouba, T, Novacek, J, Veverka, V. | | Deposit date: | 2023-01-25 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Multivalency of nucleosome recognition by LEDGF.
Nucleic Acids Res., 51, 2023
|
|
3M9Z
 
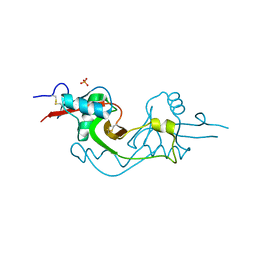 | | Crystal Structure of extracellular domain of mouse NKR-P1A | | Descriptor: | Killer cell lectin-like receptor subfamily B member 1A, PHOSPHATE ION | | Authors: | Kolenko, P, Rozbesky, D, Bezouska, K, Hasek, J, Dohnalek, J. | | Deposit date: | 2010-03-23 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular architecture of mouse activating NKR-P1 receptors.
J.Struct.Biol., 175, 2011
|
|
5K7Y
 
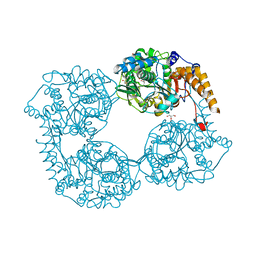 | | Crystal structure of enzyme in purine metabolism | | Descriptor: | Cytosolic purine 5'-nucleotidase, GLYCEROL | | Authors: | Skerlova, J, Hnizda, A, Pachl, P, Rezacova, P. | | Deposit date: | 2016-05-27 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Oligomeric interface modulation causes misregulation of purine 5 -nucleotidase in relapsed leukemia.
Bmc Biol., 14, 2016
|
|
5L4Z
 
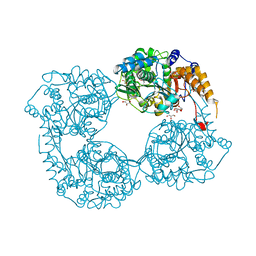 | |
6EMO
 
 | | Solution structure of the LEDGF/p75 IBD - JPO2 (aa 1-32) complex | | Descriptor: | PC4 and SFRS1-interacting protein,LEDGF/p75 IBD-JPO2 M1 | | Authors: | Veverka, V. | | Deposit date: | 2017-10-03 | | Release date: | 2018-07-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Affinity switching of the LEDGF/p75 IBD interactome is governed by kinase-dependent phosphorylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EMP
 
 | | Solution structure of the LEDGF/p75 IBD - POGZ (aa 1370-1404) complex | | Descriptor: | PC4 and SFRS1-interacting protein,Pogo transposable element with ZNF domain | | Authors: | Veverka, V. | | Deposit date: | 2017-10-03 | | Release date: | 2018-07-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Affinity switching of the LEDGF/p75 IBD interactome is governed by kinase-dependent phosphorylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EMQ
 
 | | Solution structure of the LEDGF/p75 IBD - MLL1 (aa 111-160) complex | | Descriptor: | PC4 and SFRS1-interacting protein,Histone-lysine N-methyltransferase 2A | | Authors: | Veverka, V. | | Deposit date: | 2017-10-03 | | Release date: | 2018-08-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Affinity switching of the LEDGF/p75 IBD interactome is governed by kinase-dependent phosphorylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EMR
 
 | |
3ZHO
 
 | | X-ray structure of E.coli Wrba in complex with FMN at 1.2 A resolution | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVOPROTEIN WRBA | | Authors: | Kishko, I, Lapkouski, M, Brynda, J, Kuty, M, Carey, J, Smatanova, I.K, Ettrich, R. | | Deposit date: | 2012-12-23 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | 1.2 A Resolution Crystal Structure of Escherichia Coli Wrba Holoprotein
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3CCK
 
 | | Human CD69 | | Descriptor: | CHLORIDE ION, Early activation antigen CD69 | | Authors: | Brynda, J, Vanek, O, Rezacova, P. | | Deposit date: | 2008-02-26 | | Release date: | 2008-11-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Soluble recombinant CD69 receptors optimized to have an exceptional physical and chemical stability display prolonged circulation and remain intact in the blood of mice
Febs J., 275, 2008
|
|
6TRJ
 
 | | LEDGF/p75 IBD dimer | | Descriptor: | PC4 and SFRS1-interacting protein | | Authors: | Kugler, M, Brynda, J. | | Deposit date: | 2019-12-19 | | Release date: | 2020-09-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Fine-tuning of the LEDGF/p75 interaction network by dimerization
Structure
|
|
8PC5
 
 | | H3K36me3 nucleosome-LEDGF/p75 PWWP domain complex | | Descriptor: | Histone H2A, Histone H2B 1.1, Histone H3, ... | | Authors: | Koutna, E, Kouba, T, Veverka, V. | | Deposit date: | 2023-06-09 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Multivalency of nucleosome recognition by LEDGF.
Nucleic Acids Res., 51, 2023
|
|
8PEO
 
 | | H3K36me2 nucleosome-LEDGF/p75 PWWP domain complex | | Descriptor: | (2R)-2-amino-3-(2-dimethylaminoethylsulfanyl)propanoic acid, Histone H2A, Histone H2B 1.1, ... | | Authors: | Koutna, E, Kouba, T, Veverka, V. | | Deposit date: | 2023-06-14 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Multivalency of nucleosome recognition by LEDGF.
Nucleic Acids Res., 51, 2023
|
|
