1YLO
 
 | |
1Z6M
 
 | | Structure of Conserved Protein of Unknown Function from Enterococcus faecalis V583 | | Descriptor: | PHOSPHATE ION, conserved hypothetical protein | | Authors: | Nocek, B.P, Li, H, Collart, F, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-03-22 | | Release date: | 2005-05-03 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of a conserved hypothetical protein from Enterococcus faecalis V583
To be Published
|
|
1Y9K
 
 | | IAA acetyltransferase from Bacillus cereus ATCC 14579 | | Descriptor: | IAA acetyltransferase | | Authors: | Nocek, B.P, Osipiuk, J, Li, H, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-12-15 | | Release date: | 2005-02-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | A crystal structure of IAA acetyltransferase from Bacillus cereus
To be Published
|
|
1Y0K
 
 | | Structure of Protein of Unknown Function PA4535 from Pseudomonas aeruginosa strain PAO1, Monooxygenase Superfamily | | Descriptor: | hypothetical protein PA4535 | | Authors: | Nocek, B.P, Evdokimova, E, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-11-15 | | Release date: | 2005-01-18 | | Last modified: | 2014-11-26 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | 1.75 A Crystal Structure of the Hypothetical Protein Pa4535 from Pseudomonas Aeruginosa
To be Published
|
|
3IC1
 
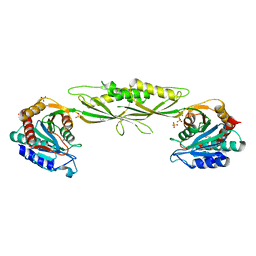 | | Crystal structure of zinc-bound succinyl-diaminopimelate desuccinylase from Haemophilus influenzae | | Descriptor: | GLYCEROL, SULFATE ION, Succinyl-diaminopimelate desuccinylase, ... | | Authors: | Nocek, B.P, Gillner, D.M, Holz, R.C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-17 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for catalysis by the mono- and dimetalated forms of the dapE-encoded N-succinyl-L,L-diaminopimelic acid desuccinylase.
J.Mol.Biol., 397, 2010
|
|
3ISZ
 
 | | Crystal structure of mono-zinc form of succinyl-diaminopimelate desuccinylase from Haemophilus influenzae | | Descriptor: | SULFATE ION, Succinyl-diaminopimelate desuccinylase, ZINC ION | | Authors: | Nocek, B.P, Gillner, D.M, Holz, R.C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-08-27 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for catalysis by the mono- and dimetalated forms of the dapE-encoded N-succinyl-L,L-diaminopimelic acid desuccinylase.
J.Mol.Biol., 397, 2010
|
|
4ZO2
 
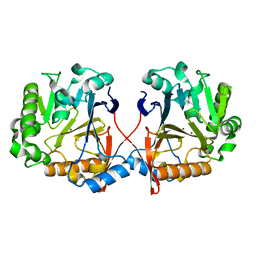 | | AidC, a Dizinc Quorum-Quenching Lactonase | | Descriptor: | Acylhomoserine lactonase, ZINC ION | | Authors: | Mascarenhas, R, Thomas, P.W, Wu, C.-X, Nocek, B.P, Hoang, Q, Fast, W, Liu, D. | | Deposit date: | 2015-05-05 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Structural and Biochemical Characterization of AidC, a Quorum-Quenching Lactonase with Atypical Selectivity.
Biochemistry, 54, 2015
|
|
4ZO3
 
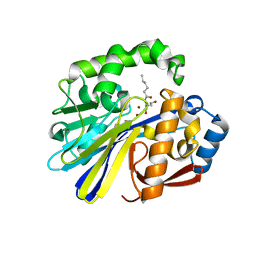 | | AidC, a Dizinc Quorum-Quenching Lactonase, in complex with a product N-hexnoyl-L-homoserine | | Descriptor: | Acylhomoserine lactonase, N-hexanoyl-L-homoserine, ZINC ION | | Authors: | Mascarenhas, R, Thomas, P.W, Wu, C.-X, Nocek, B.P, Hoang, Q, Fast, W, Liu, D. | | Deposit date: | 2015-05-05 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural and Biochemical Characterization of AidC, a Quorum-Quenching Lactonase with Atypical Selectivity.
Biochemistry, 54, 2015
|
|
2CFC
 
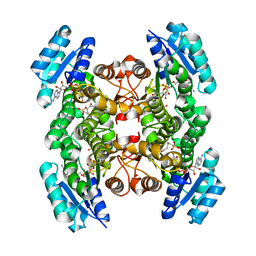 | | structural basis for stereo selectivity in the (R)- and (S)- hydroxypropylethane thiosulfonate dehydrogenases | | Descriptor: | (2-[2-KETOPROPYLTHIO]ETHANESULFONATE, 2-(R)-HYDROXYPROPYL-COM DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Krishnakumar, A.M, Nocek, B.P, Clark, D.D, Ensign, S.A, Peters, J.W. | | Deposit date: | 2006-02-19 | | Release date: | 2006-07-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Stereoselectivity in the (R)-and (S)-Hydroxypropylthioethanesulfonate Dehydrogenases.
Biochemistry, 45, 2006
|
|
1WKM
 
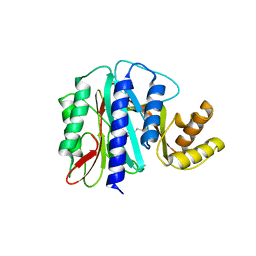 | | THE PRODUCT BOUND FORM OF THE MN(II)LOADED METHIONINE AMINOPEPTIDASE FROM HYPERTHERMOPHILE PYROCOCCUS FURIOSUS | | Descriptor: | MANGANESE (II) ION, METHIONINE, Methionine aminopeptidase | | Authors: | Copik, A.J, Nocek, B.P, Jang, S.B, Swierczek, S.I, Ruebush, S, Meng, L, D'souza, V.M, Peters, J.W, Bennett, B, Holz, R.C. | | Deposit date: | 2004-06-01 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | EPR and X-ray crystallographic characterization of the product-bound form of the MnII-loaded methionyl aminopeptidase from Pyrococcus furiosus
Biochemistry, 44, 2005
|
|
5SVB
 
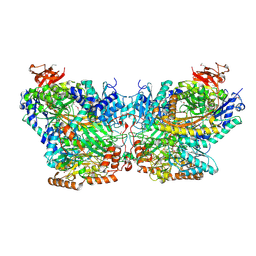 | | Mechanism of ATP-Dependent Acetone Carboxylation, Acetone Carboxylase AMP bound structure | | Descriptor: | ADENOSINE MONOPHOSPHATE, Acetone carboxylase alpha subunit, Acetone carboxylase beta subunit, ... | | Authors: | Eilers, B.J, Mus, F, Alleman, A.B, Kabasakal, B.V, Murray, J.W, Nocek, B.P, Dubois, J.L, Peters, J.W. | | Deposit date: | 2016-08-05 | | Release date: | 2017-08-09 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.645 Å) | | Cite: | Structural Basis for the Mechanism of ATP-Dependent Acetone Carboxylation.
Sci Rep, 7, 2017
|
|
5SVC
 
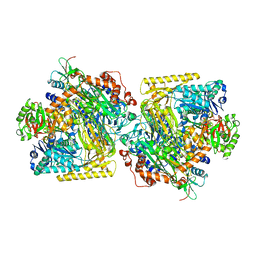 | | Mechanism of ATP-Dependent Acetone Carboxylation, Acetone Carboxylase nucleotide-free structure | | Descriptor: | Acetone carboxylase alpha subunit, Acetone carboxylase beta subunit, Acetone carboxylase gamma subunit, ... | | Authors: | Eilers, B.J, Mus, F, Alleman, A.B, Kabasakal, B.V, Murray, J.W, Nocek, B.P, Dubois, J.L, Peters, J.W. | | Deposit date: | 2016-08-05 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for the Mechanism of ATP-Dependent Acetone Carboxylation.
Sci Rep, 7, 2017
|
|
6AQE
 
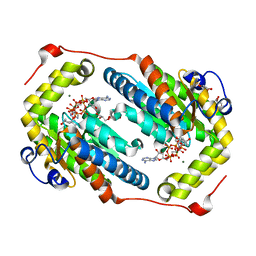 | | Crystal structure of PPK2 in complex with Mg ATP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Nocek, B, Joachimiak, A, Yakunin, A. | | Deposit date: | 2017-08-19 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Structural Insights into Substrate Selectivity and Activity of Bacterial Polyphosphate Kinases
Acs Catalysis, 8, 2018
|
|
6Y4M
 
 | | Structure of Tubulin Tyrosine Ligase in Complex with Tb111 | | Descriptor: | (2~{R})-1-methylpiperidine-2-carboxylic acid, (2~{S},4~{R})-4-azanyl-2-methyl-5-phenyl-pentanoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Gavrilyuk, J, Nocek, B, Rigol, S, Nicolaou, K.C, Stoll, V. | | Deposit date: | 2020-02-21 | | Release date: | 2021-03-31 | | Last modified: | 2022-11-23 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Tubulysin Analogues, Linker-Drugs, and Antibody-Drug Conjugates, Insights into Structure-Activity Relationships, and Tubulysin-Tubulin Binding Derived from X-ray Crystallographic Analysis.
J.Org.Chem., 86, 2021
|
|
6Y4N
 
 | | Structure of Tubulin Tyrosine Ligase in Complex with Tb116 | | Descriptor: | (2~{R})-1-methylpiperidine-2-carboxylic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Gavrilyuk, J, Nocek, B, Rigol, S, Nicolaou, K.C, Stoll, V. | | Deposit date: | 2020-02-21 | | Release date: | 2021-03-31 | | Last modified: | 2022-11-23 | | Method: | X-RAY DIFFRACTION (2.852 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Tubulysin Analogues, Linker-Drugs, and Antibody-Drug Conjugates, Insights into Structure-Activity Relationships, and Tubulysin-Tubulin Binding Derived from X-ray Crystallographic Analysis.
J.Org.Chem., 86, 2021
|
|
7O62
 
 | |
7KMA
 
 | | Crystal structure of eif2Balpha with a ligand. | | Descriptor: | 6-O-phosphono-alpha-D-mannopyranose, ACETATE ION, Translation initiation factor eIF-2B subunit alpha | | Authors: | Nocek, B, Hao, Q, Remarcik, C, Stoll, V, Wong, Y, Sidrauski, C. | | Deposit date: | 2020-11-02 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Sugar phosphate activation of the stress sensor eIF2B.
Nat Commun, 12, 2021
|
|
7KMF
 
 | | Sugar phosphate activation of the stress sensor eIF2B | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, Translation initiation factor eIF-2B subunit alpha, Translation initiation factor eIF-2B subunit beta, ... | | Authors: | Nocek, B, Hao, Q, Wong, Y, Stoll, V, Sidrauski, C. | | Deposit date: | 2020-11-02 | | Release date: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Sugar phosphate activation of the stress sensor eIF2B.
Nat Commun, 12, 2021
|
|
4XA9
 
 | | Crystal structure of the complex between the N-terminal domain of RavJ and LegL1 from Legionella pneumophila str. Philadelphia | | Descriptor: | Gala protein type 1, 3 or 4, Uncharacterized protein | | Authors: | Stogios, P.J, Cuff, M.E, Nocek, B, Evdokimova, E, Di Leo, R, Yim, V, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-12-13 | | Release date: | 2015-01-28 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Diverse mechanisms of metaeffector activity in an intracellular bacterial pathogen, Legionella pneumophila.
Mol. Syst. Biol., 12, 2016
|
|
4ZMH
 
 | | Crystal structure of a five-domain GH115 alpha-Glucuronidase from the Marine Bacterium Saccharophagus degradans 2-40T | | Descriptor: | ACETATE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Nocek, B, Cui, H, Wang, W, Savchenko, A. | | Deposit date: | 2015-05-04 | | Release date: | 2016-05-04 | | Last modified: | 2016-07-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Biochemical and Structural Characterization of a Five-domain GH115 alpha-Glucuronidase from the Marine Bacterium Saccharophagus degradans 2-40T.
J.Biol.Chem., 291, 2016
|
|
5M45
 
 | | Structure of Acetone Carboxylase purified from Xanthobacter autotrophicus | | Descriptor: | 3,6,9,12,15-PENTAOXAHEPTADECAN-1-OL, ACETATE ION, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Kabasakal, B.V, Wells, J.N, Nwaobi, B.C, Eilers, B.J, Peters, J.W, Murray, J.W. | | Deposit date: | 2016-10-18 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural Basis for the Mechanism of ATP-Dependent Acetone Carboxylation.
Sci Rep, 7, 2017
|
|
4RXI
 
 | | Structure of C-terminal domain of uncharacterized protein from Legionella pneumophila | | Descriptor: | hypothetical protein lpg0944 | | Authors: | Cuff, M, Nocek, B, Evdokimova, E, Egorova, O, Joachimiak, A, Ensminger, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-12-11 | | Release date: | 2015-05-06 | | Last modified: | 2017-01-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Diverse mechanisms of metaeffector activity in an intracellular bacterial pathogen, Legionella pneumophila.
Mol Syst Biol, 12, 2016
|
|
4RXV
 
 | | The crystal structure of the N-terminal fragment of uncharacterized protein from Legionella pneumophila | | Descriptor: | hypothetical protein lpg0944 | | Authors: | Nocek, B, Cuff, M, Evdokimova, E, Egorova, O, Joachimiak, A, Ensminger, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-12-12 | | Release date: | 2015-04-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.099 Å) | | Cite: | Diverse mechanisms of metaeffector activity in an intracellular bacterial pathogen, Legionella pneumophila.
Mol Syst Biol, 12, 2016
|
|
4PVA
 
 | | Crystal structure of GH62 hydrolase from thermophilic fungus Scytalidium thermophilum | | Descriptor: | GH62 hydrolase, GLYCEROL, PHOSPHATE ION | | Authors: | Nocek, B, Kaur, A.P, Xu, X, Cui, H, Savchenko, A. | | Deposit date: | 2014-03-15 | | Release date: | 2014-11-19 | | Last modified: | 2015-05-06 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Functional and structural diversity in GH62 alpha-L-arabinofuranosidases from the thermophilic fungus Scytalidium thermophilum.
Microb Biotechnol, 8, 2015
|
|
5DGG
 
 | | Central domain of uncharacterized Lpg1148 protein from Legionella pneumophila | | Descriptor: | CHLORIDE ION, Uncharacterized protein | | Authors: | Osipiuk, J, Evdokimova, E, Yim, V, Joachimiak, A, Ensminger, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-08-27 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Diverse mechanisms of metaeffector activity in an intracellular bacterial pathogen, Legionella pneumophila.
Mol. Syst. Biol., 12, 2016
|
|
