3B49
 
 | |
7M5H
 
 | | Crystal structure of conserved protein from Enterococcus faecalis V583 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Nucleoside 2-deoxyribosyltransferase | | Authors: | Nocek, B, Wu, R, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2021-03-23 | | Release date: | 2021-04-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of conserved protein from Enterococcus faecalis V583
To Be Published
|
|
6DZG
 
 | |
1MOK
 
 | | NADPH DEPENDENT 2-KETOPROPYL COENZYME M OXIDOREDUCTASE/CARBOXYLASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, orf3 | | Authors: | Nocek, B, Jang, S.B, Jeong, M.S, Clark, D.D, Ensign, S.A, Peters, J.W. | | Deposit date: | 2002-09-09 | | Release date: | 2002-11-27 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for CO2 Fixation by a Novel Member of the Disulfide Oxidoreductase Family of Enzymes,
2-Ketopropyl Coenzyme M Oxidoreductase/Carboxylase
Biochemistry, 41, 2002
|
|
1MO9
 
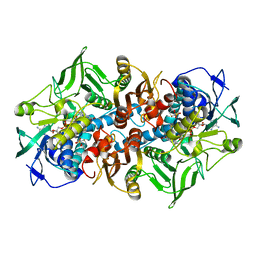 | | NADPH DEPENDENT 2-KETOPROPYL COENZYME M OXIDOREDUCTASE/CARBOXYLASE COMPLEXED WITH 2-KETOPROPYL COENZYME M | | Descriptor: | (2-[2-KETOPROPYLTHIO]ETHANESULFONATE, FLAVIN-ADENINE DINUCLEOTIDE, orf3 | | Authors: | Nocek, B, Jang, S.B, Jeong, M.S, Clark, D.D, Ensign, S.A, Peters, J.W. | | Deposit date: | 2002-09-08 | | Release date: | 2002-11-27 | | Last modified: | 2012-05-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis for CO2 Fixation by a Novel Member of the Disulfide Oxidoreductase Family of Enzymes,
2-Ketopropyl Coenzyme M Oxidoreductase/Carboxylase
Biochemistry, 41, 2002
|
|
5TGF
 
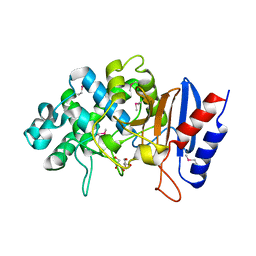 | | Crystal structure of putative beta-lactamase from Bacteroides dorei DSM 17855 | | Descriptor: | CALCIUM ION, GLYCEROL, Uncharacterized protein | | Authors: | Nocek, B, Hatzos-Skintges, C, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-09-27 | | Release date: | 2016-10-26 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of a beta-lactamase from Bacteroides dorei DSM 17855
To Be Published
|
|
4R7U
 
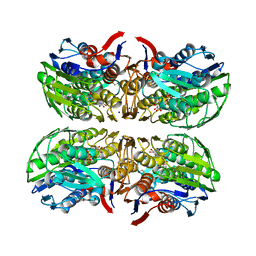 | | Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Vibrio cholerae in complex with substrate UDP-N-acetylglucosamine and the drug fosfomycin | | Descriptor: | SODIUM ION, TETRAETHYLENE GLYCOL, UDP-N-acetylglucosamine 1-carboxyvinyltransferase, ... | | Authors: | Nocek, B, Maltseva, N, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-28 | | Release date: | 2014-09-24 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Vibrio cholerae in complex with substrate UDP-N-acetylglucosamine and the drug fosfomycin
To be Published
|
|
4RW0
 
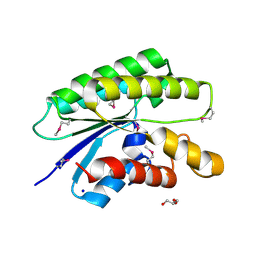 | | Crystal structure of a member of the lipolytic protein G-D-S-L family from Veillonella parvula DSM 2008 | | Descriptor: | GLYCEROL, Lipolytic protein G-D-S-L family, SODIUM ION | | Authors: | Nocek, B, Hatzos-Skintges, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-11-30 | | Release date: | 2015-01-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a member of the lipolytic protein G-D-S-L family from Veillonella parvula DSM 2008
To be Published
|
|
5INT
 
 | | Crystal structure of the C-terminal Domain of Coenzyme A biosynthesis bifunctional protein CoaBC | | Descriptor: | Phosphopantothenate--cysteine ligase | | Authors: | Nocek, B, Zhou, M, Grimshaw, S, Kim, Y, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-03-07 | | Release date: | 2016-04-06 | | Last modified: | 2018-09-19 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the C-terminal Domain of Coenzyme A biosynthesis bifunctional protein CoaBC
To Be Published
|
|
4H2K
 
 | | Crystal structure of the catalytic domain of succinyl-diaminopimelate desuccinylase from Haemophilus influenzae | | Descriptor: | Succinyl-diaminopimelate desuccinylase, ZINC ION | | Authors: | Nocek, B, Jedrzejczak, R, Makowska-Grzyska, M, Starus, A, Holz, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-09-12 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The dimerization domain in DapE enzymes is required for catalysis.
Plos One, 9, 2014
|
|
5IBZ
 
 | | Crystal structure of a novel cyclase (pfam04199). | | Descriptor: | ACETYLPHOSPHATE, TRIETHYLENE GLYCOL, Uncharacterized protein | | Authors: | Nocek, B, Skarina, T, Brown, G, Joachimiak, A, Savchenko, A, Yakunin, A. | | Deposit date: | 2016-02-22 | | Release date: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (1.611 Å) | | Cite: | Crystal structure of a novel cyclase (pfam04199).
To Be Published
|
|
5JYB
 
 | | Crystal structure of 3 mutant of Ba3275 (S116A, E243A, H313A), the member of S66 family of serine peptidases | | Descriptor: | 1,2-ETHANEDIOL, 2-BUTANOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Nocek, B, Jedrzejczak, R, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-13 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.647 Å) | | Cite: | Crystal structure of 3 mutant of Ba3275 (S116A, E243A, H313A), the member of S66 family of serine peptidases
To Be Published
|
|
5K99
 
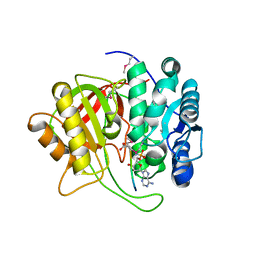 | | Crystal structure of microcin immunity protein MccF from Bacillus anthracis in complex with McC | | Descriptor: | Microcin C, Microcin C7 self-immunity protein mccF | | Authors: | Nocek, B, Kulikovsky, A, Severinov, K, Dubiley, S, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-31 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of microcin immunity protein MccF from Bacillus anthracis in complex with McC
To Be Published
|
|
5KJP
 
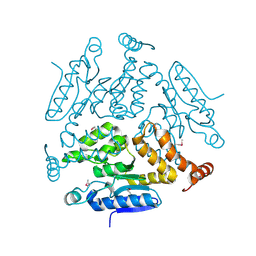 | |
5KMY
 
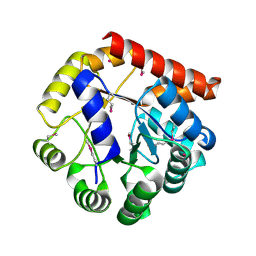 | | Crystal structure of tryptophan synthase subunit alpha from Legionella pneumophila str. Paris | | Descriptor: | Tryptophan synthase alpha chain | | Authors: | Nocek, B, Hatzos-Skintges, C, Endres, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-06-27 | | Release date: | 2016-08-03 | | Last modified: | 2017-02-08 | | Method: | X-RAY DIFFRACTION (1.908 Å) | | Cite: | Crystal structure of tryptophan synthase subunit alpha from Legionella pneumophila str. Paris
To Be Published
|
|
5DPO
 
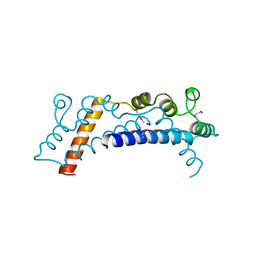 | |
5DYF
 
 | | The crystal structure of Aminopeptidase N in complex with N-benzyl-1,2-diaminoethylphosphonic acid | | Descriptor: | Aminopeptidase N, GLYCEROL, IMIDAZOLE, ... | | Authors: | Nocek, B, Joachimiak, A, Vassiliou, S, Berlicki, L, Mucha, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-09-24 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.854 Å) | | Cite: | The crystal structure of Aminopeptidase N in complex with N-benzyl-1,2-diaminoethylphosphonic acid
To Be Published
|
|
5F5X
 
 | |
5F64
 
 | | Putative positive transcription regulator (sensor EvgS) from Shigella flexneri | | Descriptor: | Positive transcription regulator EvgA | | Authors: | Nocek, B, Osipiuk, J, Mulligan, R, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-05 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Putative positive transcription regulator (sensor EvgS) from Shigella flexneri.
to be published
|
|
5F1Y
 
 | |
5F9M
 
 | | Crystal structure of native B3275, member of MccF family of enzymes | | Descriptor: | MccC family protein | | Authors: | Nocek, B, Severinov, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-09 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Crystal structure of native B3275, member of MccF family of enzymes.
To Be Published
|
|
5FCD
 
 | | Crystal structure of MccD protein | | Descriptor: | CHLORIDE ION, MccD, UNK-UNK-UNK-MSE-UNK, ... | | Authors: | Nocek, B, Jedrzejczak, R, Anderson, W.F, Severinov, K, Dubiley, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-15 | | Release date: | 2015-12-30 | | Last modified: | 2017-02-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of MccD protein.
To Be Published
|
|
5FD8
 
 | |
3S9X
 
 | | High resolution crystal structure of ASCH domain from Lactobacillus crispatus JV V101 | | Descriptor: | ASCH domain, CHLORIDE ION | | Authors: | Nocek, B, Xu, X, Cui, H, Jedrzejczak, R, Edwards, A, Savchenko, A, Mabbutt, B.C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-06-02 | | Release date: | 2011-07-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High resolution crystal structure of ASCH domain from Lactobacillus crispatus JV V101
TO BE PUBLISHED
|
|
3S52
 
 | | Crystal structure of a putative fumarylacetoacetate hydrolase family protein from Yersinia pestis CO92 | | Descriptor: | CHLORIDE ION, Putative fumarylacetoacetate hydrolase family protein, SULFATE ION | | Authors: | Nocek, B, Zhou, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-05-20 | | Release date: | 2011-06-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.012 Å) | | Cite: | Crystal structure of a putative fumarylacetoacetate hydrolase family protein from Yersinia pestis CO92
TO BE PUBLISHED
|
|
