2V5L
 
 | | Structures of the Open and Closed State of Trypanosomal Triosephosphate Isomerase: as Observed in a New Crystal Form: Implications for the Reaction Mechanism | | Descriptor: | SULFATE ION, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Noble, M.E.M, Zeelen, J.P, Wierenga, R.K. | | Deposit date: | 2007-07-06 | | Release date: | 2007-07-31 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of the Open and Closed State of Trypanosomal Triosephosphate Isomerase: As Observed in a New Crystal Form: Implications for the Reaction Mechanism
Proteins: Struct.,Funct., Genet., 16, 1993
|
|
6TIM
 
 | |
1VIN
 
 | |
8BYL
 
 | | Cryo-EM structure of SKP1-SKP2-CKS1 from the SCFSKP2 E3 ligase complex | | Descriptor: | Cyclin-dependent kinase inhibitor 1B, Cyclin-dependent kinases regulatory subunit 1, S-phase kinase-associated protein 1, ... | | Authors: | Rowland, R.J, Salamina, M, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2022-12-13 | | Release date: | 2023-06-28 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of SKP1-SKP2-CKS1 in complex with CDK2-cyclin A-p27KIP1.
Sci Rep, 13, 2023
|
|
6Q4E
 
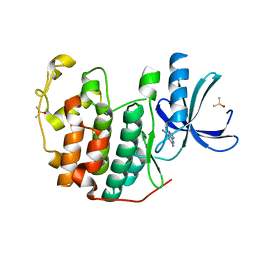 | | CDK2 in complex with FragLite33 | | Descriptor: | 6-iodanyl-7~{H}-purin-2-amine, Cyclin-dependent kinase 2, DIMETHYL SULFOXIDE | | Authors: | Wood, D.J, Martin, M.P, Noble, M.E.M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | FragLites-Minimal, Halogenated Fragments Displaying Pharmacophore Doublets. An Efficient Approach to Druggability Assessment and Hit Generation.
J.Med.Chem., 62, 2019
|
|
6Q4K
 
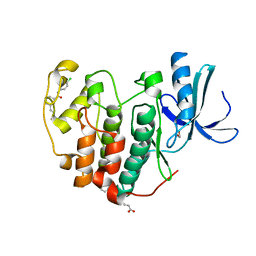 | | CDK2 in complex with FragLite38 | | Descriptor: | (~{E})-3-[3-[(4-chlorophenyl)carbamoyl]phenyl]prop-2-enoic acid, 1,2-ETHANEDIOL, Cyclin-dependent kinase 2 | | Authors: | Wood, D.J, Martin, M.P, Noble, M.E.M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | FragLites-Minimal, Halogenated Fragments Displaying Pharmacophore Doublets. An Efficient Approach to Druggability Assessment and Hit Generation.
J.Med.Chem., 62, 2019
|
|
6Q3F
 
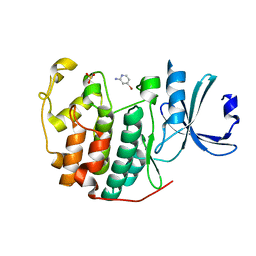 | | CDK2 in complex with FragLite2 | | Descriptor: | 4-bromanylpyridin-2-amine, Cyclin-dependent kinase 2 | | Authors: | Wood, D.J, Martin, M.P, Noble, M.E.M. | | Deposit date: | 2018-12-04 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | FragLites-Minimal, Halogenated Fragments Displaying Pharmacophore Doublets. An Efficient Approach to Druggability Assessment and Hit Generation.
J.Med.Chem., 62, 2019
|
|
6Q4D
 
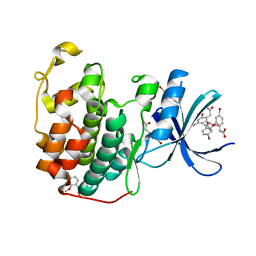 | | CDK2 in complex with FragLite31 | | Descriptor: | 2-(4-bromanyl-2-methoxy-phenyl)ethanoic acid, Cyclin-dependent kinase 2 | | Authors: | Wood, D.J, Martin, M.P, Noble, M.E.M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | FragLites-Minimal, Halogenated Fragments Displaying Pharmacophore Doublets. An Efficient Approach to Druggability Assessment and Hit Generation.
J.Med.Chem., 62, 2019
|
|
6SG4
 
 | | Structure of CDK2/cyclin A M246Q, S247EN | | Descriptor: | Cyclin-A2, Cyclin-dependent kinase 2 | | Authors: | Salamina, M, Basle, A, Massa, B, Noble, M.E.M, Endicott, J.A. | | Deposit date: | 2019-08-02 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discriminative SKP2 Interactions with CDK-Cyclin Complexes Support a Cyclin A-Specific Role in p27KIP1 Degradation.
J.Mol.Biol., 433, 2021
|
|
5LRQ
 
 | | BRD4 in complex with ERK5 inhibitor XMD8-92 | | Descriptor: | 2-{[2-ethoxy-4-(4-hydroxypiperidin-1-yl)phenyl]amino}-5,11-dimethyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain-containing protein 4 | | Authors: | Martin, M.P, Noble, M.E.M. | | Deposit date: | 2016-08-19 | | Release date: | 2017-08-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of a novel orally bioavailable ERK5 inhibitor with selectivity over p38 alpha and BRD4.
Eur.J.Med.Chem., 178, 2019
|
|
5N53
 
 | | Crystal structure of human 3-phosphoglycerate dehydrogenase in complex with N-(3-chloro-4-methoxyphenyl) acetamide | | Descriptor: | D-3-phosphoglycerate dehydrogenase, ~{N}-(3-chloranyl-4-methoxy-phenyl)ethanamide | | Authors: | Unterlass, J.E, Basle, A, Blackburn, T.J, Tucker, J, Cano, C, Noble, M.E.M, Curtin, N.J. | | Deposit date: | 2017-02-12 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Validating and enabling phosphoglycerate dehydrogenase (PHGDH) as a target for fragment-based drug discovery in PHGDH-amplified breast cancer.
Oncotarget, 9, 2018
|
|
1OW6
 
 | | Paxillin LD4 motif bound to the Focal Adhesion Targeting (FAT) domain of the Focal Adhesion Kinase | | Descriptor: | Focal adhesion kinase 1, Paxillin | | Authors: | Hoellerer, M.K, Noble, M.E.M, Labesse, G, Campbell, I.D, Werner, J.M, Arold, S.T. | | Deposit date: | 2003-03-28 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular Recognition of Paxillin LD motifs
by the Focal Adhesion Targeting Domain
Structure, 11, 2003
|
|
2IW9
 
 | | STRUCTURE OF HUMAN THR160-PHOSPHO CDK2-CYCLIN A COMPLEXED WITH A BISANILINOPYRIMIDINE INHIBITOR | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN-A2, MAGNESIUM ION, ... | | Authors: | Pratt, D.J, Bentley, J, Jewsbury, P, Boyle, F.T, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2006-06-27 | | Release date: | 2006-09-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dissecting the Determinants of Cyclin-Dependent Kinase 2 and Cyclin-Dependent Kinase 4 Inhibitor Selectivity.
J.Med.Chem., 49, 2006
|
|
2IW8
 
 | | STRUCTURE OF HUMAN THR160-PHOSPHO CDK2-CYCLIN A F82H-L83V-H84D MUTANT WITH AN O6-CYCLOHEXYLMETHYLGUANINE INHIBITOR | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN-A2, MONOTHIOGLYCEROL, ... | | Authors: | Pratt, D.J, Bentley, J, Jewsbury, P, Boyle, F.T, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2006-06-27 | | Release date: | 2006-09-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dissecting the Determinants of Cyclin-Dependent Kinase 2 and Cyclin-Dependent Kinase 4 Inhibitor Selectivity.
J.Med.Chem., 49, 2006
|
|
2IW6
 
 | | STRUCTURE OF HUMAN THR160-PHOSPHO CDK2-CYCLIN A COMPLEXED WITH A BISANILINOPYRIMIDINE INHIBITOR | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN-A2, MAGNESIUM ION, ... | | Authors: | Pratt, D.J, Bentley, J, Jewsbury, P, Boyle, F.T, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2006-06-26 | | Release date: | 2006-09-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dissecting the Determinants of Cyclin-Dependent Kinase 2 and Cyclin-Dependent Kinase 4 Inhibitor Selectivity.
J.Med.Chem., 49, 2006
|
|
2JCQ
 
 | | The hyaluronan binding domain of murine CD44 in a Type A complex with an HA 8-mer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD44 ANTIGEN, GLYCEROL | | Authors: | Banerji, S, Wright, A.J, Noble, M.E.M, Mahoney, D.J, Campbell, I.D, Day, A.J, Jackson, D.G. | | Deposit date: | 2007-01-03 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structures of the Cd44-Hyaluronan Complex Provide Insight Into a Fundamental Carbohydrate-Protein Interaction.
Nat.Struct.Mol.Biol., 14, 2008
|
|
2JCR
 
 | | The hyaluronan binding domain of murine CD44 in a Type B complex with an HA 8-mer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD44 ANTIGEN, GLYCEROL | | Authors: | Banerji, S, Wright, A.J, Noble, M.E.M, Mahoney, D.J, Campbell, I.D, Day, A.J, Jackson, D.G. | | Deposit date: | 2007-01-03 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the Cd44-Hyaluronan Complex Provide Insight Into a Fundamental Carbohydrate-Protein Interaction.
Nat.Struct.Mol.Biol., 14, 2008
|
|
2JCP
 
 | | The hyaluronan binding domain of murine CD44 | | Descriptor: | CD44 ANTIGEN | | Authors: | Banerji, S, Wright, A.J, Noble, M.E.M, Mahoney, D.J, Campbell, I.D, Day, A.J, Jackson, D.G. | | Deposit date: | 2007-01-03 | | Release date: | 2007-01-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of the Cd44-Hyaluronan Complex Provide Insight Into a Fundamental Carbohydrate-Protein Interaction.
Nat.Struct.Mol.Biol., 14, 2008
|
|
1E2T
 
 | | Arylamine N-acetyltransferase (NAT) from Salmonella typhimurium | | Descriptor: | N-HYDROXYARYLAMINE O-ACETYLTRANSFERASE | | Authors: | Sinclair, J.C, Sandy, J, Delgoda, R, Sim, E, Noble, M.E.M. | | Deposit date: | 2000-05-24 | | Release date: | 2000-07-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Arylamine N-Acetyltransferase Reveals a Catalytic Triad
Nat.Struct.Biol., 7, 2000
|
|
1Z96
 
 | | Crystal structure of the Mud1 UBA domain | | Descriptor: | UBA-domain protein mud1 | | Authors: | Trempe, J.-F, Brown, N.R, Lowe, E.D, Noble, M.E.M, Gordon, C, Campbell, I.D, Johnson, L.N, Endicott, J.A. | | Deposit date: | 2005-03-31 | | Release date: | 2005-10-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of Lys48-linked polyubiquitin chain recognition by the Mud1 UBA domain
Embo J., 24, 2005
|
|
2BWF
 
 | | Crystal structure of the UBL domain of Dsk2 from S. cerevisiae | | Descriptor: | FORMIC ACID, UBIQUITIN-LIKE PROTEIN DSK2 | | Authors: | Lowe, E.D, Hasan, N, Trempe, J.-F, Fonso, L, Noble, M.E.M, Endicott, J.A, Johnson, L.N, Brown, N.R. | | Deposit date: | 2005-07-13 | | Release date: | 2006-01-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structures of the Dsk2 Ubl and Uba Domains and Their Complex.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2C1N
 
 | | Molecular basis for the recognition of phosphorylated and phosphoacetylated histone H3 by 14-3-3 | | Descriptor: | 14-3-3 PROTEIN ZETA/DELTA, HISTONE H3 ACETYLPHOSPHOPEPTIDE | | Authors: | Welburn, J.P.I, Macdonald, N, Noble, M.E.M, Nguyen, A, Yaffe, M.B, Clynes, D, Moggs, J.G, Orphanides, G, Thomson, S, Edmunds, J.W, Clayton, A.L, Endicott, J.A, Mahadevan, L.C. | | Deposit date: | 2005-09-16 | | Release date: | 2005-11-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Basis for the Recognition of Phosphorylated and Phosphoacetylated Histone H3 by 14-3-3.
Mol.Cell, 20, 2005
|
|
2C1J
 
 | | Molecular basis for the recognition of phosphorylated and phosphoacetylated histone H3 by 14-3-3 | | Descriptor: | 14-3-3 PROTEIN ZETA/DELTA, HISTONE H3 ACETYLPHOSPHOPEPTIDE | | Authors: | Welburn, J.P.I, Macdonald, N, Noble, M.E.M, Nguyen, A, Yaffe, M.B, Clynes, D, Moggs, J.G, Orphanides, G, Thomson, S, Edmunds, J.W, Clayton, A.L, Endicott, J.A, Mahadevan, L.C. | | Deposit date: | 2005-09-15 | | Release date: | 2005-11-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Basis for the Recognition of Phosphorylated and Phosphoacetylated Histone H3 by 14-3-3.
Mol.Cell, 20, 2005
|
|
2BWB
 
 | | Crystal structure of the UBA domain of Dsk2 from S. cerevisiae | | Descriptor: | UBIQUITIN-LIKE PROTEIN DSK2 | | Authors: | Lowe, E.D, Hasan, N, Trempe, J.-F, Fonso, L, Noble, M.E.M, Endicott, J.A, Johnson, L.N, Brown, N.R. | | Deposit date: | 2005-07-13 | | Release date: | 2006-01-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of the Dsk2 Ubl and Uba Domains and Their Complex.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2BWE
 
 | | The crystal structure of the complex between the UBA and UBL domains of Dsk2 | | Descriptor: | DSK2 | | Authors: | Lowe, E.D, Hasan, N, Trempe, J.-F, Fonso, L, Noble, M.E.M, Endicott, J.A, Johnson, L.N, Brown, N.R. | | Deposit date: | 2005-07-13 | | Release date: | 2006-01-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the Dsk2 Ubl and Uba Domains and Their Complex.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
