1D8L
 
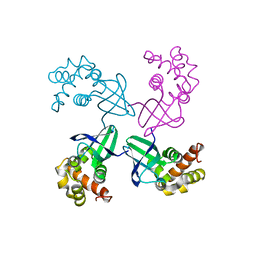 | | E. COLI HOLLIDAY JUNCTION BINDING PROTEIN RUVA NH2 REGION LACKING DOMAIN III | | Descriptor: | PROTEIN (HOLLIDAY JUNCTION DNA HELICASE RUVA) | | Authors: | Nishino, T, Iwasaki, H, Kataoka, M, Ariyoshi, M, Fujita, T, Shinagawa, H, Morikawa, K. | | Deposit date: | 1999-10-25 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Modulation of RuvB function by the mobile domain III of the Holliday junction recognition protein RuvA.
J.Mol.Biol., 298, 2000
|
|
4YTZ
 
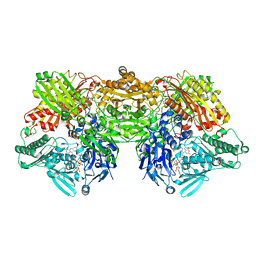 | | Rat xanthine oxidoreductase, C-terminal deletion protein variant, crystal grown without dithiothreitol | | Descriptor: | BICARBONATE ION, CALCIUM ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Nishino, T, Okamoto, K, Kawaguchi, Y, Matsumura, T, Eger, B.T, Pai, E.F, Nishino, T. | | Deposit date: | 2015-03-18 | | Release date: | 2015-04-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The C-terminal peptide plays a role in the formation of an intermediate form during the transition between xanthine dehydrogenase and xanthine oxidase
Febs J., 282, 2015
|
|
4YTY
 
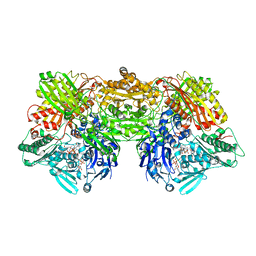 | | Structure of rat xanthine oxidoreductase, C535A/C992R/C1324S, NADH bound form | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, BICARBONATE ION, CALCIUM ION, ... | | Authors: | Nishino, T, Okamoto, K, Kawaguchi, Y, Matsumura, T, Eger, B.T, Pai, E.F. | | Deposit date: | 2015-03-18 | | Release date: | 2015-04-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The C-terminal peptide plays a role in the formation of an intermediate form during the transition between xanthine dehydrogenase and xanthine oxidase.
Febs J., 282, 2015
|
|
4YSW
 
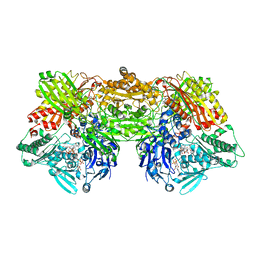 | | Structure of rat xanthine oxidoreductase, C-terminal deletion protein variant, NADH bound form | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, BICARBONATE ION, CALCIUM ION, ... | | Authors: | Nishino, T, Okamoto, K, Kawaguchi, Y, Matsumura, T, Eger, B.T, Pai, E.F. | | Deposit date: | 2015-03-17 | | Release date: | 2015-04-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The C-terminal peptide plays a role in the formation of an intermediate form during the transition between xanthine dehydrogenase and xanthine oxidase.
Febs J., 282, 2015
|
|
3B0C
 
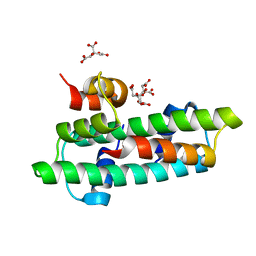 | | Crystal structure of the chicken CENP-T histone fold/CENP-W complex, crystal form I | | Descriptor: | CITRIC ACID, Centromere protein T, Centromere protein W | | Authors: | Nishino, T, Takeuchi, K, Gascoigne, K.E, Suzuki, A, Hori, T, Oyama, T, Morikawa, K, Cheeseman, I.M, Fukagawa, T. | | Deposit date: | 2011-06-08 | | Release date: | 2012-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | CENP-T-W-S-X Forms a Unique Centromeric Chromatin Structure with a Histone-like Fold.
Cell(Cambridge,Mass.), 148, 2012
|
|
3B0B
 
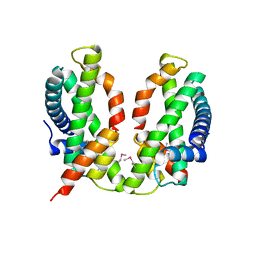 | | Crystal structure of the chicken CENP-S/CENP-X complex | | Descriptor: | Centromere protein S, Centromere protein X | | Authors: | Nishino, T, Takeuchi, K, Gascoigne, K.E, Suzuki, A, Hori, T, Oyama, T, Morikawa, K, Cheeseman, I.M, Fukagawa, T. | | Deposit date: | 2011-06-08 | | Release date: | 2012-03-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | CENP-T-W-S-X Forms a Unique Centromeric Chromatin Structure with a Histone-like Fold.
Cell(Cambridge,Mass.), 148, 2012
|
|
1WYG
 
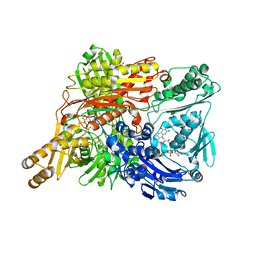 | | Crystal Structure of a Rat Xanthine Dehydrogenase Triple Mutant (C535A, C992R and C1324S) | | Descriptor: | 2-HYDROXYBENZOIC ACID, ACETIC ACID, CALCIUM ION, ... | | Authors: | Nishino, T, Okamoto, K, Kawaguchi, Y, Hori, H, Matsumura, T, Eger, B.T, Pai, E.F, Nishino, T. | | Deposit date: | 2005-02-14 | | Release date: | 2005-05-31 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanism of the Conversion of Xanthine Dehydrogenase to Xanthine Oxidase: IDENTIFICATION OF THE TWO CYSTEINE DISULFIDE BONDS AND CRYSTAL STRUCTURE OF A NON-CONVERTIBLE RAT LIVER XANTHINE DEHYDROGENASE MUTANT
J.Biol.Chem., 280, 2005
|
|
1GEF
 
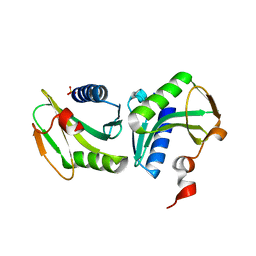 | | Crystal structure of the archaeal holliday junction resolvase HJC | | Descriptor: | HOLLIDAY JUNCTION RESOLVASE, SULFATE ION | | Authors: | Nishino, T, Komori, K, Tsuchiya, D, Ishino, Y, Morikawa, K. | | Deposit date: | 2000-11-08 | | Release date: | 2001-03-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the archaeal holliday junction resolvase Hjc and implications for DNA recognition.
Structure, 9, 2001
|
|
1IPI
 
 | |
1HJP
 
 | | HOLLIDAY JUNCTION BINDING PROTEIN RUVA FROM E. COLI | | Descriptor: | RUVA | | Authors: | Nishino, T, Ariyoshi, M, Iwasaki, H, Shinagawa, H, Morikawa, K. | | Deposit date: | 1997-08-21 | | Release date: | 1998-02-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Functional Analyses of the Domain Structure in the Holliday Junction Binding Protein Ruva
Structure, 6, 1998
|
|
1J23
 
 | | Crystal structure of archaeal XPF/Mus81 homolog, Hef from Pyrococcus furiosus, nuclease domain | | Descriptor: | ATP-dependent RNA helicase, putative | | Authors: | Nishino, T, Komori, K, Ishino, Y, Morikawa, K. | | Deposit date: | 2002-12-25 | | Release date: | 2003-04-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | X-Ray and Biochemical Anatomy of an Archaeal XPF/Rad1/Mus81 Family Nuclease. Similarity between Its Endonuclease Domain and Restriction Enzymes
Structure, 11, 2003
|
|
1J25
 
 | | Crystal structure of archaeal XPF/Mus81 homolog, Hef from Pyrococcus furiosus, nuclease domain, Mn cocrystal | | Descriptor: | ATP-dependent RNA helicase, putative, MANGANESE (II) ION | | Authors: | Nishino, T, Komori, K, Ishino, Y, Morikawa, K. | | Deposit date: | 2002-12-25 | | Release date: | 2003-04-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | X-Ray and Biochemical Anatomy of an Archaeal XPF/Rad1/Mus81 Family Nuclease. Similarity between Its Endonuclease Domain and Restriction Enzymes
Structure, 11, 2003
|
|
1J24
 
 | | Crystal structure of archaeal XPF/Mus81 homolog, Hef from Pyrococcus furiosus, nuclease domain, Ca cocrystal | | Descriptor: | ATP-dependent RNA helicase, putative, CALCIUM ION | | Authors: | Nishino, T, Komori, K, Ishino, Y, Morikawa, K. | | Deposit date: | 2002-12-25 | | Release date: | 2003-04-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | X-Ray and Biochemical Anatomy of an Archaeal XPF/Rad1/Mus81 Family Nuclease. Similarity between Its Endonuclease Domain and Restriction Enzymes
Structure, 11, 2003
|
|
1J22
 
 | | Crystal structure of archaeal XPF/Mus81 homolog, Hef from Pyrococcus furiosus, nuclease domain, selenomet derivative | | Descriptor: | ATP-dependent RNA helicase, putative | | Authors: | Nishino, T, Komori, K, Ishino, Y, Morikawa, K. | | Deposit date: | 2002-12-25 | | Release date: | 2003-04-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray and Biochemical Anatomy of an Archaeal XPF/Rad1/Mus81 Family Nuclease. Similarity between Its Endonuclease Domain and Restriction Enzymes
Structure, 11, 2003
|
|
3VH6
 
 | | Crystal structure of the chicken CENP-T histone fold/CENP-W/CENP-S/CENP-X heterotetrameric complex, crystal form II | | Descriptor: | CENP-S, CENP-T, CENP-W, ... | | Authors: | Nishino, T, Takeuchi, K, Gascoigne, K.E, Suzuki, A, Hori, T, Oyama, T, Morikawa, K, Cheeseman, I.M, Fukagawa, T. | | Deposit date: | 2011-08-23 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.351 Å) | | Cite: | CENP-T-W-S-X Forms a Unique Centromeric Chromatin Structure with a Histone-like Fold
Cell(Cambridge,Mass.), 148, 2012
|
|
3VH5
 
 | | Crystal structure of the chicken CENP-T histone fold/CENP-W/CENP-S/CENP-X heterotetrameric complex, crystal form I | | Descriptor: | CENP-S, CENP-T, CENP-W, ... | | Authors: | Nishino, T, Takeuchi, K, Gascoigne, K.E, Suzuki, A, Hori, T, Oyama, T, Morikawa, K, Cheeseman, I.M, Fukagawa, T. | | Deposit date: | 2011-08-23 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | CENP-T-W-S-X Forms a Unique Centromeric Chromatin Structure with a Histone-like Fold
Cell(Cambridge,Mass.), 148, 2012
|
|
3VZ9
 
 | |
3VZA
 
 | |
1WP9
 
 | | Crystal structure of Pyrococcus furiosus Hef helicase domain | | Descriptor: | ATP-dependent RNA helicase, putative, PHOSPHATE ION | | Authors: | Nishino, T, Komori, K, Tsuchiya, D, Ishino, Y, Morikawa, K. | | Deposit date: | 2004-08-31 | | Release date: | 2005-02-01 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure and Functional Implications of Pyrococcus furiosus Hef Helicase Domain Involved in Branched DNA Processing
Structure, 13, 2005
|
|
1X2I
 
 | | Crystal Structure Of Archaeal Xpf/Mus81 Homolog, Hef From Pyrococcus Furiosus, Helix-hairpin-helix Domain | | Descriptor: | Hef helicase/nuclease | | Authors: | Nishino, T, Komori, K, Ishino, Y, Morikawa, K. | | Deposit date: | 2005-04-24 | | Release date: | 2005-09-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and Functional Analyses of an Archaeal XPF/Rad1/Mus81 Nuclease: Asymmetric DNA Binding and Cleavage Mechanisms
STRUCTURE, 13, 2005
|
|
3B0D
 
 | | Crystal structure of the chicken CENP-T histone fold/CENP-W complex, crystal form II | | Descriptor: | CITRIC ACID, Centromere protein T, Centromere protein W | | Authors: | Nishino, T, Takeuchi, K, Gascoigne, K.E, Suzuki, A, Hori, T, Oyama, T, Morikawa, K, Cheeseman, I.M, Fukagawa, T. | | Deposit date: | 2011-06-08 | | Release date: | 2012-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | CENP-T-W-S-X Forms a Unique Centromeric Chromatin Structure with a Histone-like Fold.
Cell(Cambridge,Mass.), 148, 2012
|
|
7DA2
 
 | | The crystal structure of the chicken FANCM-MHF complex | | Descriptor: | Centromere protein S, Centromere protein X, Fanconi anemia group M protein | | Authors: | Nishino, T, Ito, S. | | Deposit date: | 2020-10-14 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural analysis of the chicken FANCM-MHF complex and its stability.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
3BDJ
 
 | | Crystal Structure of Bovine Milk Xanthine Dehydrogenase with a Covalently Bound Oxipurinol Inhibitor | | Descriptor: | CALCIUM ION, CARBONATE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Eger, B.T, Okamoto, K, Nishino, T, Pai, E.F, Nishino, T. | | Deposit date: | 2007-11-14 | | Release date: | 2008-11-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of inhibition of xanthine oxidoreductase by allopurinol: crystal structure of reduced bovine milk xanthine oxidoreductase bound with oxipurinol.
Nucleosides Nucleotides Nucleic Acids, 27, 2008
|
|
1FO4
 
 | | CRYSTAL STRUCTURE OF XANTHINE DEHYDROGENASE ISOLATED FROM BOVINE MILK | | Descriptor: | 2-HYDROXYBENZOIC ACID, CALCIUM ION, DIOXOTHIOMOLYBDENUM(VI) ION, ... | | Authors: | Enroth, C, Eger, B.T, Okamoto, K, Nishino, T, Nishino, T, Pai, E.F. | | Deposit date: | 2000-08-24 | | Release date: | 2000-10-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of bovine milk xanthine dehydrogenase and xanthine oxidase: structure-based mechanism of conversion.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1FIQ
 
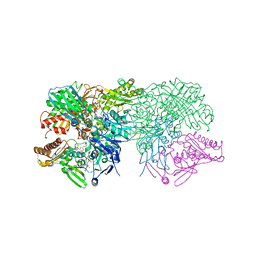 | | CRYSTAL STRUCTURE OF XANTHINE OXIDASE FROM BOVINE MILK | | Descriptor: | 2-HYDROXYBENZOIC ACID, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Enroth, C, Eger, B.T, Okamoto, K, Nishino, T, Nishino, T, Pai, E.F. | | Deposit date: | 2000-08-04 | | Release date: | 2000-10-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of bovine milk xanthine dehydrogenase and xanthine oxidase: structure-based mechanism of conversion.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
