5EGM
 
 | | Development of a novel tricyclic class of potent and selective FIXa inhibitors | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 2-chloranyl-~{N}-[(7~{S})-2-methyl-7-phenyl-10-(1~{H}-1,2,3,4-tetrazol-5-yl)-8,9-dihydro-6~{H}-pyrido[1,2-a]indol-7-yl]-4-(1,2,4-triazol-4-yl)benzamide, Coagulation factor IX, ... | | Authors: | Meng, D, Andre, P, Bateman, T.J, Berger, R, Chen, Y, Desai, K, Dewnani, S, Ellsworth, K, Feng, D, Geissler, W.M, Guo, L, Hruza, A, Jian, T, Li, H, Parker, D.L, Reichert, P, Sherer, E.C, Smith, C.J, Sonatore, L.M, Tschirret-Guth, R, Wu, J, Xu, J, Zhang, T, Campeau, L, Orr, R, Poirier, M, McCabe-Dunn, j, Araki, K, Nishimura, T, Sakurada, I, Hirabayashi, T, Wood, H.B. | | Deposit date: | 2015-10-27 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.841 Å) | | Cite: | Development of a novel tricyclic class of potent and selective FIXa inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
5TNO
 
 | | Discovery of novel aminobenzisoxazole derivatives as orally available factor IXa inhibitors | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Coagulation factor IX, SODIUM ION, ... | | Authors: | Sakurada, I, Endo, T, Hikita, K, Hirabayashi, T, Hosaka, Y, Kato, Y, Maeda, Y, Matsumoto, S, Mizuno, T, Nagasue, H, Nishimura, T, Shimada, S, Shinozaki, M, Taguchi, K, Takeuchi, K, Yokoyama, T, Hruza, A, Reichert, P, Zhang, T, Wood, H.B, Nakao, K, Furusako, S. | | Deposit date: | 2016-10-14 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Discovery of novel aminobenzisoxazole derivatives as orally available factor IXa inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5TNT
 
 | | Discovery of novel aminobenzisoxazole derivatives as orally available factor IXa inhibitors | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Coagulation factor IX, N-[(1S,4S,7R)-2-(3-amino-4-chloro[1,2]oxazolo[5,4-c]pyridin-7-yl)-2-azabicyclo[2.2.1]heptan-7-yl]-2-chloro-4-(3-methyl-1H-1,2,4-triazol-1-yl)benzamide, ... | | Authors: | Sakurada, I, Endo, T, Hikita, K, Hirabayashi, T, Hosaka, Y, Kato, Y, Maeda, Y, Matsumoto, S, Mizuno, T, Nagasue, A, Nishimura, T, Shimada, S, Shinozaki, M, Taguchi, K, Takeuchi, K, Yokoyama, T, Hruza, A, Reichert, P, Zhang, T, Wood, H.B, Nakao, K, Furusako, S. | | Deposit date: | 2016-10-14 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of novel aminobenzisoxazole derivatives as orally available factor IXa inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
6L0V
 
 | | Structure of RLD2 BRX domain bound to LZY3 CCL motif | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NGR2, ... | | Authors: | Hirano, Y, Futrutani, M, Nishimura, T, Taniguchi, M, Morita, M.T, Hakoshima, T. | | Deposit date: | 2019-09-27 | | Release date: | 2020-02-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.347 Å) | | Cite: | Polar recruitment of RLD by LAZY1-like protein during gravity signaling in root branch angle control.
Nat Commun, 11, 2020
|
|
6L0W
 
 | | Structure of RLD2 BRX domain bound to LZY3 CCL motif | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, NGR2, ... | | Authors: | Hirano, Y, Futrutani, M, Nishimura, T, Taniguchi, M, Morita, M.T, Hakoshima, T. | | Deposit date: | 2019-09-27 | | Release date: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Polar recruitment of RLD by LAZY1-like protein during gravity signaling in root branch angle control.
Nat Commun, 11, 2020
|
|
1V4T
 
 | | Crystal structure of human glucokinase | | Descriptor: | SODIUM ION, SULFATE ION, glucokinase isoform 2 | | Authors: | Kamata, K, Mitsuya, M, Nishimura, T, Eiki, J, Nagata, Y. | | Deposit date: | 2003-11-19 | | Release date: | 2004-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis for allosteric regulation of the monomeric allosteric enzyme human glucokinase
Structure, 12, 2004
|
|
1V4S
 
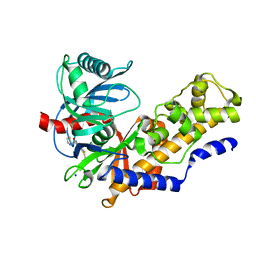 | | Crystal structure of human glucokinase | | Descriptor: | 2-AMINO-4-FLUORO-5-[(1-METHYL-1H-IMIDAZOL-2-YL)SULFANYL]-N-(1,3-THIAZOL-2-YL)BENZAMIDE, SODIUM ION, alpha-D-glucopyranose, ... | | Authors: | Kamata, K, Mitsuya, M, Nishimura, T, Eiki, J, Nagata, Y. | | Deposit date: | 2003-11-19 | | Release date: | 2004-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for allosteric regulation of the monomeric allosteric enzyme human glucokinase
Structure, 12, 2004
|
|
3FR0
 
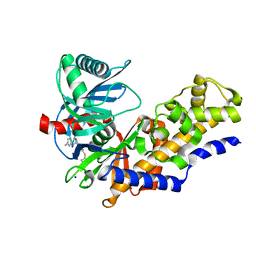 | | Human glucokinase in complex with 2-amino benzamide activator | | Descriptor: | 2-amino-N-(4-methyl-1,3-thiazol-2-yl)-5-[(4-methyl-4H-1,2,4-triazol-3-yl)sulfanyl]benzamide, Glucokinase, SODIUM ION, ... | | Authors: | Kamata, K. | | Deposit date: | 2009-01-08 | | Release date: | 2009-02-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of novel and potent 2-amino benzamide derivatives as allosteric glucokinase activators
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6FDL
 
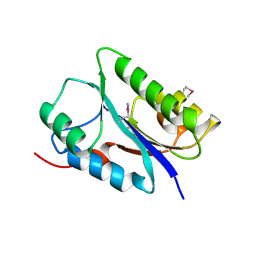 | | Crystal structure of the NYN domain of human MARF1 | | Descriptor: | Meiosis regulator and mRNA stability factor 1 | | Authors: | Jinek, M, Brandmann, T. | | Deposit date: | 2017-12-26 | | Release date: | 2018-11-07 | | Last modified: | 2018-12-26 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Human MARF1 is an endoribonuclease that interacts with the DCP1:2 decapping complex and degrades target mRNAs.
Nucleic Acids Res., 46, 2018
|
|
3VEM
 
 | | Structural basis of transcriptional gene silencing mediated by Arabidopsis MOM1 | | Descriptor: | Helicase protein MOM1 | | Authors: | Nishikura, T, Petty, T.J, Halazonetis, T, Paszkowski, J, Thore, S. | | Deposit date: | 2012-01-09 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis of Transcriptional Gene Silencing Mediated by Arabidopsis MOM1.
PLOS GENET., 8, 2012
|
|
1CVZ
 
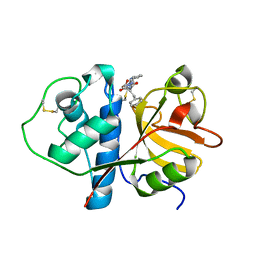 | | CRYSTAL STRUCTURE ANALYSIS OF PAPAIN WITH CLIK148(CATHEPSIN L SPECIFIC INHIBITOR) | | Descriptor: | N1-(1-DIMETHYLCARBAMOYL-2-PHENYL-ETHYL)-2-OXO-N4-(2-PYRIDIN-2-YL-ETHYL)-SUCCINAMIDE, PAPAIN | | Authors: | Tsuge, H. | | Deposit date: | 1999-08-24 | | Release date: | 2000-08-30 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition mechanism of cathepsin L-specific inhibitors based on the crystal structure of papain-CLIK148 complex.
Biochem.Biophys.Res.Commun., 266, 1999
|
|
7EBV
 
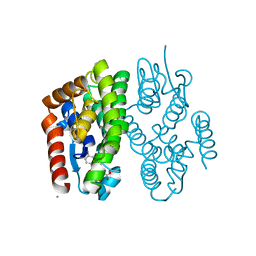 | | Crystal structure of Aedes aegypti Noppera-bo, glutathione S-transferase epsilon 8, in luteolin- and glutathione-bound form | | Descriptor: | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4H-chromen-4-one, CALCIUM ION, GLUTATHIONE, ... | | Authors: | Inaba, K, Koiwai, K, Senda, M, Senda, T, Niwa, R. | | Deposit date: | 2021-03-11 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular action of larvicidal flavonoids on ecdysteroidogenic glutathione S-transferase Noppera-bo in Aedes aegypti.
Bmc Biol., 20, 2022
|
|
7EBT
 
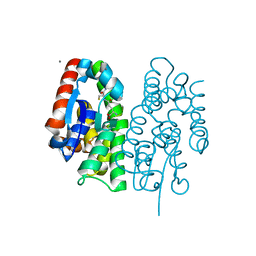 | | Crystal structure of Aedes aegypti Noppera-bo, glutathione S-transferase epsilon 8, in glutathione-bound form | | Descriptor: | CALCIUM ION, GLUTATHIONE, Glutathione transferase | | Authors: | Inaba, K, Koiwai, K, Senda, M, Senda, T, Niwa, R. | | Deposit date: | 2021-03-11 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Molecular action of larvicidal flavonoids on ecdysteroidogenic glutathione S-transferase Noppera-bo in Aedes aegypti.
Bmc Biol., 20, 2022
|
|
7EBW
 
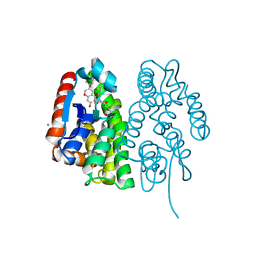 | | Crystal structure of Aedes aegypti Noppera-bo, glutathione S-transferase epsilon 8, in desmethylglycitein and glutathione-bound form | | Descriptor: | 6,7-dihydroxy-3-(4-hydroxyphenyl)-4H-chromen-4-one, CALCIUM ION, GLUTATHIONE, ... | | Authors: | Inaba, K, Koiwai, K, Senda, M, Senda, T, Niwa, R. | | Deposit date: | 2021-03-11 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Molecular action of larvicidal flavonoids on ecdysteroidogenic glutathione S-transferase Noppera-bo in Aedes aegypti.
Bmc Biol., 20, 2022
|
|
7EBU
 
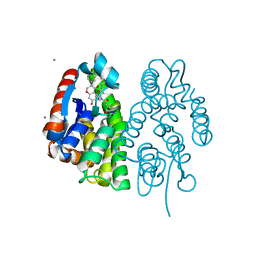 | | Crystal structure of Aedes aegypti Noppera-bo, glutathione S-transferase epsilon 8, in Daidzein- and glutathione-bound form | | Descriptor: | 7-hydroxy-3-(4-hydroxyphenyl)-4H-chromen-4-one, CALCIUM ION, GLUTATHIONE, ... | | Authors: | Inaba, K, Koiwai, K, Senda, M, Senda, T, Niwa, R. | | Deposit date: | 2021-03-11 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular action of larvicidal flavonoids on ecdysteroidogenic glutathione S-transferase Noppera-bo in Aedes aegypti.
Bmc Biol., 20, 2022
|
|
1HL8
 
 | |
1HL9
 
 | | CRYSTAL STRUCTURE OF THERMOTOGA MARITIMA ALPHA-FUCOSIDASE IN COMPLEX WITH A MECHANISM BASED INHIBITOR | | Descriptor: | 2-deoxy-2-fluoro-beta-L-fucopyranose, PUTATIVE ALPHA-L-FUCOSIDASE | | Authors: | Sulzenbacher, G, Bignon, C, Bourne, Y, Henrissat, B. | | Deposit date: | 2003-03-14 | | Release date: | 2004-02-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Thermotoga Maritima {Alpha}-L-Fucosidase: Insights Into the Catalytic Mechanism and the Molecular Basis for Fucosidosis
J.Biol.Chem., 279, 2004
|
|
4ZAE
 
 | | Development of a novel class of potent and selective FIXa inhibitors | | Descriptor: | 2,6-dichloro-N-[(2R)-2-(5,6-dimethyl-1H-benzimidazol-2-yl)-2-phenylethyl]-4-(4H-1,2,4-triazol-4-yl)benzamide, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Coagulation factor IX, ... | | Authors: | Hruza, A, Reichert, P. | | Deposit date: | 2015-04-13 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Development of a novel class of potent and selective FIXa inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
3H1V
 
 | | Human glucokinase in complex with a synthetic activator | | Descriptor: | 1-({5-[4-(methylsulfonyl)phenoxy]-2-pyridin-2-yl-1H-benzimidazol-6-yl}methyl)pyrrolidine-2,5-dione, Glucokinase, SODIUM ION, ... | | Authors: | Kamata, K, Takahashi, K. | | Deposit date: | 2009-04-14 | | Release date: | 2009-10-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The design and optimization of a series of 2-(pyridin-2-yl)-1H-benzimidazole compounds as allosteric glucokinase activators.
Bioorg.Med.Chem., 17, 2009
|
|
3GOI
 
 | | Human glucokinase in complex with a synthetic activator | | Descriptor: | 2-(methylamino)-N-(4-methyl-1,3-thiazol-2-yl)-5-[(4-methyl-4H-1,2,4-triazol-3-yl)sulfanyl]benzamide, Glucokinase, alpha-D-glucopyranose | | Authors: | Kamata, K, Mitsuya, M. | | Deposit date: | 2009-03-19 | | Release date: | 2009-04-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Discovery of novel 3,6-disubstituted 2-pyridinecarboxamide derivatives as GK activators
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3ANT
 
 | | Human soluble epoxide hydrolase in complex with a synthetic inhibitor | | Descriptor: | 4-[3-(1-methylethyl)-1,2,4-oxadiazol-5-yl]-N-[(1S,2R)-2-phenylcyclopropyl]piperidine-1-carboxamide, Epoxide hydrolase 2 | | Authors: | Chiyo, N, Ishii, T, Hourai, S, Yanagi, K. | | Deposit date: | 2010-09-08 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A practical use of ligand efficiency indices out of the fragment-based approach: ligand efficiency-guided lead identification of soluble epoxide hydrolase inhibitors
J.Med.Chem., 54, 2011
|
|
3ANS
 
 | | Human soluble epoxide hydrolase in complex with a synthetic inhibitor | | Descriptor: | 4-cyano-N-[(1S,2R)-2-phenylcyclopropyl]benzamide, Epoxide hydrolase 2 | | Authors: | Chiyo, N, Ishii, T, Hourai, S, Yanagi, K. | | Deposit date: | 2010-09-08 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | A practical use of ligand efficiency indices out of the fragment-based approach: ligand efficiency-guided lead identification of soluble epoxide hydrolase inhibitors
J.Med.Chem., 54, 2011
|
|
6KPD
 
 | |
6KPB
 
 | |
6IKN
 
 | | Crystal structure of the GAS7 F-BAR domain | | Descriptor: | Growth arrest-specific protein 7 | | Authors: | Hanawa-Suetsugu, K, Itoh, Y, Kohda, D, Shimada, A, Suetsugu, S. | | Deposit date: | 2018-10-16 | | Release date: | 2019-10-16 | | Last modified: | 2019-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Phagocytosis is mediated by two-dimensional assemblies of the F-BAR protein GAS7.
Nat Commun, 10, 2019
|
|
