4QE8
 
 | | FXR with DM175 and NCoA-2 peptide | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,2-ETHANEDIOL, 4-({2-[(4-tert-butylbenzoyl)amino]benzoyl}amino)benzoic acid, ... | | Authors: | Kudlinzki, D, Merk, D, Linhard, V.L, Saxena, K, Sreeramulu, S, Nilsson, E, Dekker, N, Wissler, L, Bamberg, K, Schubert-Zsilavecz, M, Schwalbe, H. | | Deposit date: | 2014-05-15 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | FXR with DM175 and NCoA-2 peptide
To be Published
|
|
4UDB
 
 | | MR in complex with desisobutyrylciclesonide | | Descriptor: | DESISOBUYTYRYL CICLESONIDE, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Edman, K, Hogner, A, Hussein, A, Aagaard, A, Backstrom, S, Bodin, C, Wissler, L, JellesmarkJensen, T, Cavallin, A, Nilsson, E, Lepisto, M, Guallar, V. | | Deposit date: | 2014-12-09 | | Release date: | 2015-11-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Ligand Binding Mechanism in Steroid Receptors: From Conserved Plasticity to Differential Evolutionary Constraints.
Structure, 23, 2015
|
|
4UDD
 
 | | GR in complex with desisobutyrylciclesonide | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, DESISOBUYTYRYL CICLESONIDE, ... | | Authors: | Edman, K, Hogner, A, Hussein, A, Bjursell, M, Aagaard, A, Backstrom, S, Bodin, C, Wissler, L, Jellesmark-Jensen, T, Cavallin, A, Karlsson, U, Nilsson, E, Lecina, D, Takahashi, R, Grebner, C, Lepisto, M, Guallar, V. | | Deposit date: | 2014-12-09 | | Release date: | 2015-11-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ligand Binding Mechanism in Steroid Receptors: From Conserved Plasticity to Differential Evolutionary Constraints.
Structure, 23, 2015
|
|
4UDC
 
 | | GR in complex with dexamethasone | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, DEXAMETHASONE, GLUCOCORTICOID RECEPTOR, ... | | Authors: | Edman, K, Hogner, A, Hussein, A, Bjursell, M, Aagaard, A, Backstrom, S, Bodin, C, Wissler, L, Jellesmark-Jensen, T, Cavallin, A, Karlsson, U, Nilsson, E, Lecina, D, Takahashi, R, Grebner, C, Lepisto, M, Guallar, V. | | Deposit date: | 2014-12-09 | | Release date: | 2015-11-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand Binding Mechanism in Steroid Receptors: From Conserved Plasticity to Differential Evolutionary Constraints.
Structure, 23, 2015
|
|
4UDA
 
 | | MR in complex with dexamethasone | | Descriptor: | DEXAMETHASONE, GLYCEROL, MINERALOCORTICOID RECEPTOR, ... | | Authors: | Edman, K, Hogner, A, Hussein, A, Bjursell, M, Aagaard, A, Backstrom, S, Bodin, C, Wissler, L, Jellesmark-Jensen, T, Cavallin, A, Karlsson, U, Nilsson, E, Lecina, D, Takahashi, R, Grebner, C, Lepisto, M, Guallar, V. | | Deposit date: | 2014-12-09 | | Release date: | 2015-11-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Ligand Binding Mechanism in Steroid Receptors: From Conserved Plasticity to Differential Evolutionary Constraints.
Structure, 23, 2015
|
|
5FX6
 
 | | Novel inhibitors of human rhinovirus 3C protease | | Descriptor: | RHINOVIRUS 3C PROTEASE, ethyl (4R)-4-[[(2S,4S)-1-[(2S)-3-methyl-2-[(5-methyl-1,2-oxazol-3-yl)carbonylamino]butanoyl]-4-phenyl-pyrrolidin-2-yl]carbonylamino]-5-[(3S)-2-oxidanylidenepyrrolidin-3-yl]pentanoate | | Authors: | Kawatkar, S, Ek, M, Hoesch, V, Gagnon, M, Nilsson, E, Lister, T, Olsson, L, Patel, J, Yu, Q. | | Deposit date: | 2016-02-24 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Design and Structure-Activity Relationships of Novel Inhibitors of Human Rhinovirus 3C Protease.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5FX5
 
 | | Novel inhibitors of human rhinovirus 3C protease | | Descriptor: | GLYCEROL, RHINOVIRUS 3C PROTEASE, TETRAETHYLENE GLYCOL, ... | | Authors: | Kawatkar, S, Ek, M, Hoesch, V, Gagnon, M, Nilsson, E, Lister, T, Olsson, L, Patel, J, Yu, Q. | | Deposit date: | 2016-02-24 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design and Structure-Activity Relationships of Novel Inhibitors of Human Rhinovirus 3C Protease.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
2WGT
 
 | | Structure of human adenovirus serotype 37 fibre head in complex with a sialic acid derivative, O-Methyl 5-N-propaonyl-3,5-dideoxy-D- glycero-a-D-galacto-2-nonulopyranosylonic acid | | Descriptor: | 3,5-dideoxy-5-(propanoylamino)-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid, FIBER PROTEIN, ZINC ION | | Authors: | Johansson, S, Nilsson, E, Qian, W, Guilligay, D, Crepin, T, Cusack, S, Arnberg, N, Elofsson, M. | | Deposit date: | 2009-04-27 | | Release date: | 2009-11-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, Synthesis, and Evaluation of N-Acyl Modified Sialic Acids as Inhibitors of Adenoviruses Causing Epidemic Keratoconjunctivitis.
J.Med.Chem., 52, 2009
|
|
2XSM
 
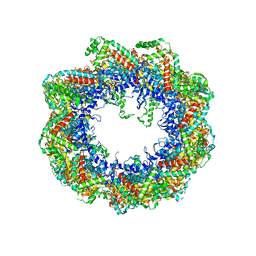 | | Crystal structure of the mammalian cytosolic chaperonin CCT in complex with tubulin | | Descriptor: | CCT | | Authors: | Munoz, I.G, Yebenes, H, Zhou, M, Mesa, P, Serna, M, Bragado-Nilsson, E, Beloso, A, Robinson, C.V, Valpuesta, J.M, Montoya, G. | | Deposit date: | 2010-10-29 | | Release date: | 2010-12-15 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Crystal Structure of the Open Conformation of the Mammalian Chaperonin Cct in Complex with Tubulin.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3BZI
 
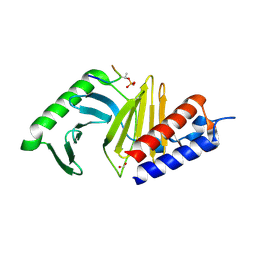 | | Molecular and structural basis of polo-like kinase 1 substrate recognition: Implications in centrosomal localization | | Descriptor: | 9 mer peptide from M-phase inducer phosphatase 3, FORMIC ACID, Serine/threonine-protein kinase PLK1 | | Authors: | Garcia-Alvarez, B, de Carcer, G, Ibanez, S, Bragado-Nilsson, E, Montoya, G. | | Deposit date: | 2008-01-18 | | Release date: | 2008-02-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular and structural basis of polo-like kinase 1 substrate recognition: Implications in centrosomal localization.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2OGQ
 
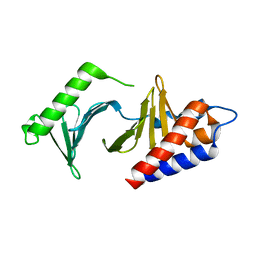 | | Molecular and structural basis of Plk1 substrate recognition: Implications in centrosomal localization | | Descriptor: | Serine/threonine-protein kinase PLK1 | | Authors: | Garcia-Alvarez, B, de Carcer, G, Ibanez, S, Bragado-Nilsson, E, Montoya, G. | | Deposit date: | 2007-01-08 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular and structural basis of polo-like kinase 1 substrate recognition: Implications in centrosomal localization.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2OJX
 
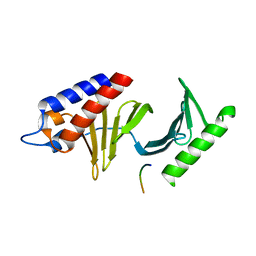 | | Molecular and structural basis of polo-like kinase 1 substrate recognition: Implications in centrosomal localization | | Descriptor: | Serine/threonine-protein kinase PLK1, Synthetic peptide | | Authors: | Garcia-Alvarez, B, de Carcer, G, Ibanez, S, Bragado-Nilsson, E, Montoya, G. | | Deposit date: | 2007-01-15 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular and structural basis of polo-like kinase 1 substrate recognition: Implications in centrosomal localization.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2WGU
 
 | | Structure of human adenovirus serotype 37 fibre head in complex with a sialic acid derivative, O-Methyl 5-N- methoxycarbonyl -3,5-dideoxy- D-glycero-a-D-galacto-2-nonulopyranosylonic acid | | Descriptor: | 3,5-dideoxy-5-[(methoxycarbonyl)amino]-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid, FIBER PROTEIN, ZINC ION | | Authors: | Johansson, S, Nilsson, E, Qian, W, Guilligay, D, Crepin, T, Cusack, S, Arnberg, N, Elofsson, M. | | Deposit date: | 2009-04-27 | | Release date: | 2009-11-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, Synthesis, and Evaluation of N-Acyl Modified Sialic Acids as Inhibitors of Adenoviruses Causing Epidemic Keratoconjunctivitis.
J.Med.Chem., 52, 2009
|
|
6VR4
 
 | |
6HL1
 
 | | Crystal Structure of Farnesoid X receptor (FXR) with bound NCoA-2 peptide and CDCA | | Descriptor: | Bile acid receptor, CHENODEOXYCHOLIC ACID, NCoA-2 peptide (Nuclear receptor coactivator 2), ... | | Authors: | Kudlinzki, D, Merk, D, Linhard, V.L, Saxena, K, Schubert-Zsilavecz, M, Schwalbe, H. | | Deposit date: | 2018-09-10 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Molecular tuning of farnesoid X receptor partial agonism.
Nat Commun, 10, 2019
|
|
6HL0
 
 | | Crystal Structure of Farnesoid X receptor (FXR) with bound NCoA-2 peptide | | Descriptor: | Bile acid receptor, NCoA-2 peptide (Nuclear receptor coactivator 2), LYS-GLU-ASN-ALA-LEU-LEU-ARG-TYR-LEU-LEU-ASP-LYS-ASP | | Authors: | Kudlinzki, D, Merk, D, Linhard, V.L, Saxena, K, Schubert-Zsilavecz, M, Schwalbe, H. | | Deposit date: | 2018-09-10 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Molecular tuning of farnesoid X receptor partial agonism.
Nat Commun, 10, 2019
|
|
4XL8
 
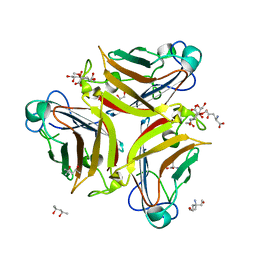 | | Crystal Structure of Human Adenovirus 52 Short Fiber Knob in Complex with 2-O-Methyl-5-N-Acetylneuraminic Acid | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-O-methyl-5-N-acetyl-alpha-D-neuraminic acid, ... | | Authors: | Stehle, T, Liaci, A.M. | | Deposit date: | 2015-01-13 | | Release date: | 2015-01-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Human Adenovirus 52 Uses Sialic Acid-containing Glycoproteins and the Coxsackie and Adenovirus Receptor for Binding to Target Cells.
Plos Pathog., 11, 2015
|
|
2JA4
 
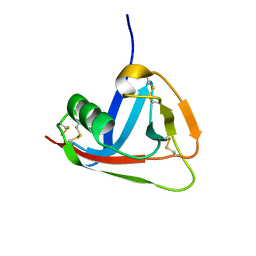 | | Crystal structure of CD5 domain III reveals the fold of a group B scavenger cysteine-rich receptor | | Descriptor: | T-CELL SURFACE GLYCOPROTEIN CD5 | | Authors: | Rodamilans, B, Munoz, I.G, Sarrias, M.R, Lozano, F, Blanco, F.J, Montoya, G. | | Deposit date: | 2006-11-21 | | Release date: | 2007-03-06 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal Structure of the Third Extracellular Domain of Cd5 Reveals the Fold of a Group B Scavenger Cysteine-Rich Receptor Domain.
J.Biol.Chem., 282, 2007
|
|
4B2T
 
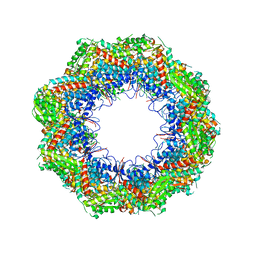 | | The crystal structures of the eukaryotic chaperonin CCT reveal its functional partitioning | | Descriptor: | T-COMPLEX PROTEIN 1 SUBUNIT ALPHA, T-COMPLEX PROTEIN 1 SUBUNIT BETA, T-COMPLEX PROTEIN 1 SUBUNIT DELTA, ... | | Authors: | Kalisman, N, Schroeder, G.F, Levitt, M. | | Deposit date: | 2012-07-17 | | Release date: | 2013-03-20 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | The Crystal Structures of the Eukaryotic Chaperonin Cct Reveal its Functional Partitioning
Structure, 21, 2013
|
|
2OTT
 
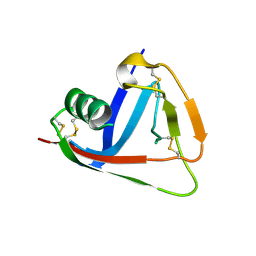 | | Crystal structure of CD5_DIII | | Descriptor: | T-cell surface glycoprotein CD5 | | Authors: | Rodamilans, B. | | Deposit date: | 2007-02-09 | | Release date: | 2007-03-13 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the third extracellular domain of CD5 reveals the fold of a group B scavenger cysteine-rich receptor domain.
J.Biol.Chem., 282, 2007
|
|
2VCW
 
 | | Complex structure of prostaglandin D2 synthase at 1.95A. | | Descriptor: | 1-PHENYL-1H-PYRAZOLE-4-CARBOXYLIC ACID, GLUTATHIONE, GLUTATHIONE-REQUIRING PROSTAGLANDIN D SYNTHASE | | Authors: | Hohwy, M, Spadola, L, Lundquist, B, von Wachenfeldt, K, Persdotter, S, Hawtin, P, Dahmen, J, Groth-Clausen, I, Folmer, R.H.A, Edman, K. | | Deposit date: | 2007-09-27 | | Release date: | 2008-04-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Novel Prostaglandin D Synthase Inhibitors Generated by Fragment-Based Drug Design.
J.Med.Chem., 51, 2008
|
|
2VCZ
 
 | | Complex structure of prostaglandin D2 synthase at 1.95A. | | Descriptor: | 3-(4-nitrophenyl)-1H-pyrazole, GLUTATHIONE, GLUTATHIONE-REQUIRING PROSTAGLANDIN D SYNTHASE | | Authors: | Hohwy, M, Spadola, L, Lundquist, B, von Wachenfeldt, K, Persdotter, S, Hawtin, P, Dahmen, J, Groth-Clausen, I, Folmer, R.H.A, Edman, K. | | Deposit date: | 2007-09-28 | | Release date: | 2008-04-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Novel Prostaglandin D Synthase Inhibitors Generated by Fragment-Based Drug Design.
J.Med.Chem., 51, 2008
|
|
2VCQ
 
 | | Complex structure of prostaglandin D2 synthase at 1.95A. | | Descriptor: | 3-phenyl-5-(1H-pyrazol-3-yl)isoxazole, GLUTATHIONE, GLUTATHIONE-REQUIRING PROSTAGLANDIN D SYNTHASE | | Authors: | Hohwy, M, Spadola, L, Lundquist, B, von Wachenfeldt, K, Persdotter, S, Hawtin, P, Dahmen, J, Groth-Clausen, I, Folmer, R.H.A, Edman, K. | | Deposit date: | 2007-09-26 | | Release date: | 2008-04-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Novel Prostaglandin D Synthase Inhibitors Generated by Fragment-Based Drug Design.
J.Med.Chem., 51, 2008
|
|
2VD0
 
 | | Complex structure of prostaglandin D2 synthase at 2.2A. | | Descriptor: | 2-{[(2E)-3-(3,4-dimethoxyphenyl)prop-2-enoyl]amino}benzoic acid, GLUTATHIONE, GLUTATHIONE-REQUIRING PROSTAGLANDIN D SYNTHASE, ... | | Authors: | Hohwy, M, Spadola, L, Lundquist, B, von Wachenfeldt, K, Persdotter, S, Hawtin, P, Dahmen, J, Groth-Clausen, I, Folmer, R.H.A, Edman, K. | | Deposit date: | 2007-09-28 | | Release date: | 2008-04-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel Prostaglandin D Synthase Inhibitors Generated by Fragment-Based Drug Design.
J.Med.Chem., 51, 2008
|
|
2VD1
 
 | | Complex structure of prostaglandin D2 synthase at 2.25A. | | Descriptor: | 4-{[4-(4-fluoro-3-methylphenyl)-1,3-thiazol-2-yl]amino}-2-hydroxybenzoic acid, GLUTATHIONE, GLUTATHIONE-REQUIRING PROSTAGLANDIN D SYNTHASE, ... | | Authors: | Hohwy, M, Spadola, L, Lundquist, B, von Wachenfeldt, K, Persdotter, S, Hawtin, P, Dahmen, J, Groth-Clausen, I, Folmer, R.H.A, Edman, K. | | Deposit date: | 2007-09-28 | | Release date: | 2008-04-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Novel Prostaglandin D Synthase Inhibitors Generated by Fragment-Based Drug Design.
J.Med.Chem., 51, 2008
|
|
