6VZC
 
 | |
17RA
 
 | |
2LV0
 
 | |
5UF3
 
 | |
1D0T
 
 | |
1D0U
 
 | |
4PDB
 
 | |
2AWQ
 
 | |
1J4Y
 
 | |
1JU7
 
 | |
1JWC
 
 | |
1KKA
 
 | |
1BGZ
 
 | |
2LA9
 
 | | NMR structure of Pseudouridine_ASL_Tyr | | Descriptor: | RNA (5'-R(*GP*GP*GP*GP*AP*CP*UP*GP*UP*AP*AP*AP*(PSU)P*CP*CP*CP*C)-3') | | Authors: | Denmon, A.P, Wang, J, Nikonowicz, E.P. | | Deposit date: | 2011-03-09 | | Release date: | 2011-08-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Conformation Effects of Base Modification on the Anticodon Stem-Loop of Bacillus subtilis tRNA(Tyr).
J.Mol.Biol., 412, 2011
|
|
2LBL
 
 | |
2LBJ
 
 | |
2LBQ
 
 | | NMR structure of i6A37_tyrASL | | Descriptor: | RNA (5'-R(*GP*GP*GP*GP*AP*CP*UP*GP*UP*AP*(6IA)P*AP*UP*CP*CP*CP*C)-3') | | Authors: | Denmon, A.P, Wang, J, Nikonowicz, E.P. | | Deposit date: | 2011-04-05 | | Release date: | 2011-08-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Conformation Effects of Base Modification on the Anticodon Stem-Loop of Bacillus subtilis tRNA(Tyr).
J.Mol.Biol., 412, 2011
|
|
2LBK
 
 | |
2LBR
 
 | |
2LAC
 
 | | NMR structure of unmodified_ASL_Tyr | | Descriptor: | RNA (5'-R(*GP*GP*GP*GP*AP*CP*UP*GP*UP*AP*AP*AP*UP*CP*CP*CP*C)-3') | | Authors: | Denmon, A.P, Wang, J, Nikonowicz, E.P. | | Deposit date: | 2011-03-10 | | Release date: | 2011-08-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Conformation Effects of Base Modification on the Anticodon Stem-Loop of Bacillus subtilis tRNA(Tyr).
J.Mol.Biol., 412, 2011
|
|
2LUN
 
 | |
2GBH
 
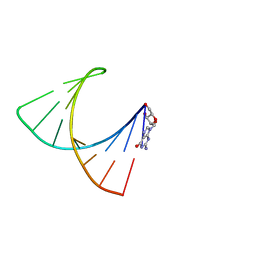 | | NMR structure of stem region of helix-35 of 23S E.coli ribosomal RNA (residues 736-760) | | Descriptor: | 5'-R(*(GMP)P*GP*GP*CP*UP*AP*AP*UP*GP*(PSU)P*UP*GP*AP*AP*AP*AP*AP*UP*UP*AP*GP*CP*CP*C)-3' | | Authors: | Bax, A, Boisbouvier, J, Bryce, D, Grishaev, A, Jaroniec, C, Miclet, E, Nikonovicz, E, O'Neil-Cabello, E, Ying, J. | | Deposit date: | 2006-03-10 | | Release date: | 2006-04-11 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Measurement of five dipolar couplings from a single 3D NMR multiplet applied to the study of RNA dynamics.
J.Am.Chem.Soc., 126, 2004
|
|
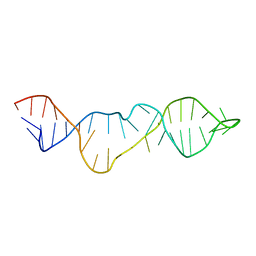
2KHY
 
 | |
1I6U
 
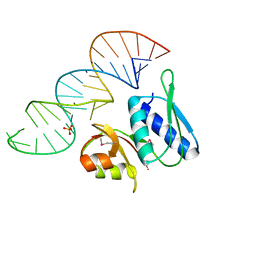 | | RNA-PROTEIN INTERACTIONS: THE CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN S8/RRNA COMPLEX FROM METHANOCOCCUS JANNASCHII | | Descriptor: | 16S RRNA FRAGMENT, 30S RIBOSOMAL PROTEIN S8P, SULFATE ION | | Authors: | Tishchenko, S, Nikulin, A, Fomenkova, N, Nevskaya, N, Nikonov, O, Dumas, P, Moine, H, Ehresmann, B, Ehresmann, C, Piendl, W, Lamzin, V, Garber, M, Nikonov, S. | | Deposit date: | 2001-03-05 | | Release date: | 2001-08-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Detailed analysis of RNA-protein interactions within the ribosomal protein S8-rRNA complex from the archaeon Methanococcus jannaschii.
J.Mol.Biol., 311, 2001
|
|
