1YZB
 
 | | Solution structure of the Josephin domain of Ataxin-3 | | Descriptor: | Machado-Joseph disease protein 1 | | Authors: | Nicastro, G, Masino, L, Menon, R.P, Knowles, P.P, McDonald, N.Q, Pastore, A. | | Deposit date: | 2005-02-28 | | Release date: | 2005-07-05 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the Josephin domain of ataxin-3: Structural determinants for molecular recognition
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
8COO
 
 | | Solution structure of Zipcode binding protein 1 (ZBP1) KH3(DD)KH4 domains in complex with N6-Methyladenosine containing RNA | | Descriptor: | Insulin-like growth factor 2 mRNA-binding protein 1, RNA_(5'-R(*(UP*CP*GP*GP*(6MZ)P*CP*U)-3') | | Authors: | Nicastro, G, Abis, G, Taylor, I.A, Ramos, A. | | Deposit date: | 2023-02-28 | | Release date: | 2024-02-07 | | Method: | SOLUTION NMR | | Cite: | Direct m6A recognition by IMP1 underlays an alternative model of target selection for non-canonical methyl-readers.
Nucleic Acids Res., 51, 2023
|
|
2BIC
 
 | | The solution structure of the recombinant elicitor protein PcF from the oomycete pathogen P. cactorum | | Descriptor: | PHYTOTOXIC PROTEIN PCF | | Authors: | Nicastro, G, Orsomando, G, Desario, F, Ferrari, E, Manconi, L, Spisni, A, Ruggieri, S. | | Deposit date: | 2005-01-20 | | Release date: | 2006-06-28 | | Last modified: | 2020-01-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Phytotoxic Protein Pcf: The First Characterized Member of the Phytophthora Pcf Toxin Family.
Protein Sci., 18, 2009
|
|
1QUW
 
 | | SOLUTION STRUCTURE OF THE THIOREDOXIN FROM BACILLUS ACIDOCALDARIUS | | Descriptor: | THIOREDOXIN | | Authors: | Nicastro, G, de Chiara, C, Pedone, E, Tato, M, Rossi, M. | | Deposit date: | 1999-07-02 | | Release date: | 2000-01-26 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a novel thioredoxin from Bacillus acidocaldarius possible determinants of protein stability.
Eur.J.Biochem., 267, 2000
|
|
4B8T
 
 | | RNA BINDING PROTEIN Solution structure of the third KH domain of KSRP in complex with the G-rich target sequence. | | Descriptor: | 5'-R(*AP*GP*GP*GP*UP)-3', KH-TYPE SPLICING REGULATORY PROTEIN | | Authors: | Nicastro, G, Garcia-Mayoral, M.F, Hollingworth, D, Kelly, G, Martin, S.R, Briata, P, Gherzi, R, Ramos, A. | | Deposit date: | 2012-08-30 | | Release date: | 2012-11-07 | | Last modified: | 2013-01-16 | | Method: | SOLUTION NMR | | Cite: | Noncanonical G Recognition Mediates Ksrp Regulation of Let-7 Biogenesis
Nat.Struct.Mol.Biol., 19, 2012
|
|
2JRI
 
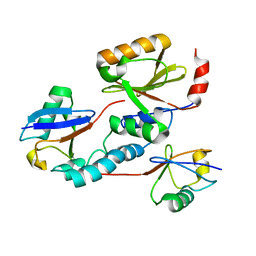 | | Solution structure of the Josephin domain of Ataxin-3 in complex with ubiquitin molecule. | | Descriptor: | Ataxin-3, UBC protein | | Authors: | Nicastro, G, Masino, L, Esposito, V, Menon, R, Pastore, A. | | Deposit date: | 2007-06-27 | | Release date: | 2008-07-01 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Understanding the plasticity of the ubiquitin-protein recognition code: the josephin domain of ataxin-3 is a diubiquitin binding motif
To be Published
|
|
7ZUD
 
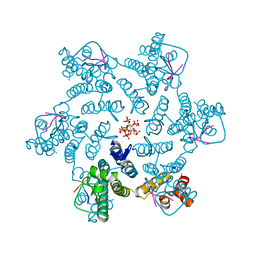 | | Crystal structure of HIV-1 capsid IP6-CPSF6 complex | | Descriptor: | Capsid protein p24, Cleavage and polyadenylation specificity factor subunit 6, INOSITOL HEXAKISPHOSPHATE | | Authors: | Nicastro, G, Taylor, I.A. | | Deposit date: | 2022-05-12 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | CP-MAS and Solution NMR Studies of Allosteric Communication in CA-assemblies of HIV-1.
J.Mol.Biol., 434, 2022
|
|
2N8M
 
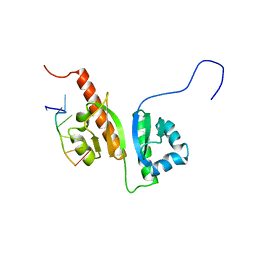 | |
2N8L
 
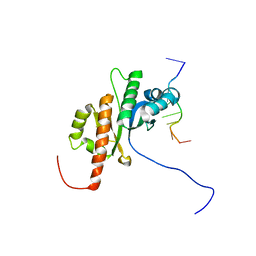 | |
1H5O
 
 | | Solution structure of Crotamine, a neurotoxin from Crotalus durissus terrificus | | Descriptor: | MYOTOXIN | | Authors: | Nicastro, G, Franzoni, L, De Chiara, C, Mancin, C.A, Giglio, J.R, Spisni, A. | | Deposit date: | 2001-05-23 | | Release date: | 2003-05-09 | | Last modified: | 2013-07-24 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Crotamine, a Na+ Channel Affecting Toxin from Crotalus Durissus Terrificus Venom
Eur.J.Biochem., 270, 2003
|
|
5M1G
 
 | |
6SAI
 
 | |
5M1H
 
 | |
6SSL
 
 | |
6SA9
 
 | |
6SSJ
 
 | |
6SSM
 
 | |
6SSK
 
 | |
