4V10
 
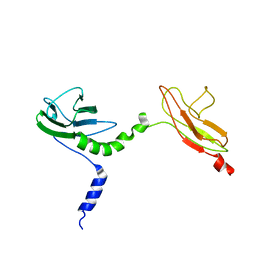 | | Skelemin Association with alpha2b,beta3 Integrin: A Structural Model | | Descriptor: | MYOMESIN-1 | | Authors: | Gorbatyuk, V, Deshmukh, L, Nguyen, K, Vinogradova, O. | | Deposit date: | 2014-09-19 | | Release date: | 2014-10-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Skelemin Association with Alphaiibbeta3 Integrin: A Structural Model.
Biochemistry, 53, 2014
|
|
3R22
 
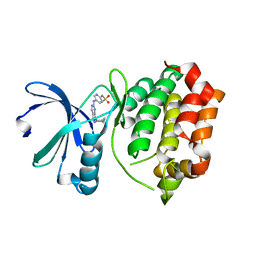 | | Design, synthesis, and biological evaluation of pyrazolopyridine-sulfonamides as potent multiple-mitotic kinase (MMK) inhibitors (Part I) | | Descriptor: | N-{5-[(1-cycloheptyl-1H-pyrazolo[3,4-d]pyrimidin-6-yl)amino]pyridin-2-yl}methanesulfonamide, Serine/threonine-protein kinase 6 | | Authors: | Zhang, L, Fan, J, Chong, J.-H, Cesana, A, Tam, B, Gilson, C, Boykin, C, Wang, D, Marcotte, D, Le Brazidec, J.-Y, Aivazian, D, Piao, J, Lundgren, K, Hong, K, Vu, K, Nguyen, K. | | Deposit date: | 2011-03-11 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design, synthesis, and biological evaluation of pyrazolopyrimidine-sulfonamides as potent multiple-mitotic kinase (MMK) inhibitors (part I).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3R21
 
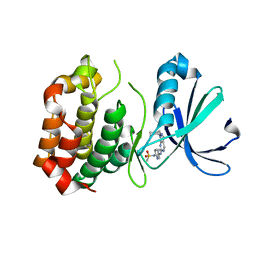 | | Design, synthesis, and biological evaluation of pyrazolopyridine-sulfonamides as potent multiple-mitotic kinase (MMK) inhibitors (Part I) | | Descriptor: | MAGNESIUM ION, N-(2-aminoethyl)-N-{5-[(1-cycloheptyl-1H-pyrazolo[3,4-d]pyrimidin-6-yl)amino]pyridin-2-yl}methanesulfonamide, Serine/threonine-protein kinase 6 | | Authors: | Zhang, L, Fan, J, Chong, J.-H, Cesena, A, Tam, B, Gilson, C, Boykin, C, Wang, D, Marcotte, D, Le Brazidec, J.-Y, Aivazian, D, Piao, J, Lundgren, K, Hong, K, Vu, K, Nguyen, K. | | Deposit date: | 2011-03-11 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design, synthesis, and biological evaluation of pyrazolopyrimidine-sulfonamides as potent multiple-mitotic kinase (MMK) inhibitors (part I).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2ML7
 
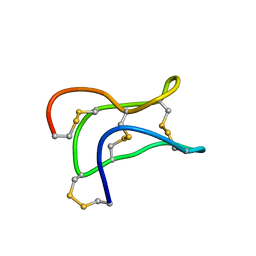 | | Ginsentides: Characterization, Structure and Application of a New Class of Highly Stable Cystine Knot Peptides in Ginseng | | Descriptor: | Specific abundant protein 3 | | Authors: | Wang, S, Nguyen, K, Luo, S, Tam, J, Yang, D. | | Deposit date: | 2014-02-20 | | Release date: | 2015-03-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Ginsentides: Characterization, Structure and Application of a New Class of Highly Stable Cystine Knot Peptides in Ginseng
To be Published
|
|
3R4V
 
 | | Structure of the phage tubulin PhuZ-GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative uncharacterized protein | | Authors: | Agard, D.A, Pogliano, J, Kraemer, J.A, Erb, M.L, Waddling, C.A, Montabana, E.A, Wang, H, Nguyen, K, Pham, S. | | Deposit date: | 2011-03-17 | | Release date: | 2012-07-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | A phage tubulin assembles dynamic filaments by an atypical mechanism to center viral DNA within the host cell.
Cell(Cambridge,Mass.), 149, 2012
|
|
3RB8
 
 | | Structure of the phage tubulin PhuZ(SeMet)-GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative uncharacterized protein | | Authors: | Agard, D.A, Pogliano, J, Kraemer, J.A, Erb, M.L, Waddling, C.A, Montabana, E.A, Wang, H, Nguyen, K, Pham, S. | | Deposit date: | 2011-03-28 | | Release date: | 2012-07-04 | | Last modified: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A phage tubulin assembles dynamic filaments by an atypical mechanism to center viral DNA within the host cell.
Cell(Cambridge,Mass.), 149, 2012
|
|
3D03
 
 | | 1.9A structure of Glycerophoshphodiesterase (GpdQ) from Enterobacter aerogenes | | Descriptor: | COBALT (II) ION, Phosphohydrolase | | Authors: | Hadler, K.S, Tanifum, E, Yip, S.H.-C, Miti, N, Guddat, L.W, Jackson, C.J, Gahan, L.R, Carr, P.D, Nguyen, K, Ollis, D.L, Hengge, A.C, Larrabee, J.A, Schenk, G. | | Deposit date: | 2008-04-30 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate-promoted formation of a catalytically competent binuclear center and regulation of reactivity in a glycerophosphodiesterase from Enterobacter aerogenes.
J.Am.Chem.Soc., 130, 2008
|
|
2XS4
 
 | | Structure of karilysin catalytic MMP domain in complex with magnesium | | Descriptor: | CHLORIDE ION, KARILYSIN PROTEASE, MAGNESIUM ION, ... | | Authors: | Cerda-Costa, N, Guevara, T, Karim, A.Y, Ksiazek, M, Nguyen, K.-A, Arolas, J.L, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2010-09-24 | | Release date: | 2010-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure of the Catalytic Domain of Tannerella Forsythia Karilysin Reveals It is a Bacterial Xenologue of Animal Matrix Metalloproteinases.
Mol.Microbiol., 79, 2011
|
|
2XS3
 
 | | Structure of karilysin catalytic MMP domain | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, KARILYSIN PROTEASE, PEPTIDE ALA-PHE-THR-SER, ... | | Authors: | Cerda-Costa, N, Guevara, T, Karim, A.Y, Ksiazek, M, Nguyen, K.-A, Arolas, J.L, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2010-09-24 | | Release date: | 2010-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Structure of the Catalytic Domain of Tannerella Forsythia Karilysin Reveals It is a Bacterial Xenologue of Animal Matrix Metalloproteinases.
Mol.Microbiol., 79, 2011
|
|
7LZQ
 
 | | Crystal structure of the BCL6 BTB domain in complex with OICR-4425 | | Descriptor: | B-cell lymphoma 6 protein, DIMETHYL SULFOXIDE, N-(3-chloropyridin-4-yl)-2-(5-methyl-4-oxo-4,5-dihydro-1H-pyrrolo[3,2-c]pyridin-1-yl)acetamide | | Authors: | Kuntz, D.A, Prive, G.G. | | Deposit date: | 2021-03-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Discovery of OICR12694: A Novel, Potent, Selective, and Orally Bioavailable BCL6 BTB Inhibitor.
Acs Med.Chem.Lett., 14, 2023
|
|
7LWG
 
 | | Crystal structure of the BCL6 BTB domain in complex with OICR-12694 | | Descriptor: | 5-{(5S)-1-[2-({3-chloro-6-[(2S)-2,4-dimethylpiperazin-1-yl]-2-fluoropyridin-4-yl}amino)-2-oxoethyl]-4-oxo-4,6,7,8-tetrahydro-1H-dipyrrolo[1,2-a:2',3'-d]pyrimidin-3-yl}-3,4-difluoro-2-hydroxybenzamide, B-cell lymphoma 6 protein, GLYCEROL, ... | | Authors: | Kuntz, D.A, Prive, G.G. | | Deposit date: | 2021-03-01 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of OICR12694: A Novel, Potent, Selective, and Orally Bioavailable BCL6 BTB Inhibitor.
Acs Med.Chem.Lett., 14, 2023
|
|
7LWE
 
 | | Crystal structure of the BCL6 BTB domain in complex with OICR-7629 | | Descriptor: | ACETATE ION, B-cell lymphoma 6 protein, CHLORIDE ION, ... | | Authors: | Kuntz, D.A, Prive, G.G. | | Deposit date: | 2021-03-01 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Discovery of OICR12694: A Novel, Potent, Selective, and Orally Bioavailable BCL6 BTB Inhibitor.
Acs Med.Chem.Lett., 14, 2023
|
|
7LWF
 
 | | Crystal structure of the BCL6 BTB domain in complex with OICR-9320 | | Descriptor: | B-cell lymphoma 6 protein, GLYCEROL, N-(3-chloropyridin-4-yl)-2-[5-(3-cyano-4-hydroxyphenyl)-3-methyl-4-oxo-3,4-dihydro-7H-pyrrolo[2,3-d]pyrimidin-7-yl]acetamide | | Authors: | Kuntz, D.A, Prive, G.G. | | Deposit date: | 2021-03-01 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Discovery of OICR12694: A Novel, Potent, Selective, and Orally Bioavailable BCL6 BTB Inhibitor.
Acs Med.Chem.Lett., 14, 2023
|
|
7LZS
 
 | | Crystal structure of the BCL6 BTB domain in complex with OICR-11029 | | Descriptor: | 3-chloro-5-{7-[2-({5-chloro-2-[(3S)-3-methylmorpholin-4-yl]pyridin-4-yl}amino)-2-oxoethyl]-3-methyl-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl}-2-hydroxybenzamide, B-cell lymphoma 6 protein, DIMETHYL SULFOXIDE, ... | | Authors: | Kuntz, D.A, Prive, G.G. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Discovery of OICR12694: A Novel, Potent, Selective, and Orally Bioavailable BCL6 BTB Inhibitor.
Acs Med.Chem.Lett., 14, 2023
|
|
3KHQ
 
 | | Crystal structure of murine Ig-beta (CD79b) in the monomeric form | | Descriptor: | B-cell antigen receptor complex-associated protein beta chain, CITRATE ANION, GLUTATHIONE, ... | | Authors: | Radaev, S, Sun, P.D. | | Deposit date: | 2009-10-30 | | Release date: | 2010-08-25 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Studies of Igalphabeta and Its Assembly with the B Cell Antigen Receptor.
Structure, 18, 2010
|
|
3KG5
 
 | | Crystal structure of human Ig-beta homodimer | | Descriptor: | B-cell antigen receptor complex-associated protein beta chain | | Authors: | Radaev, S, Sun, P.D. | | Deposit date: | 2009-10-28 | | Release date: | 2010-08-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and Functional Studies of Igalphabeta and Its Assembly with the B Cell Antigen Receptor.
Structure, 18, 2010
|
|
3KHO
 
 | | Crystal structure of murine Ig-beta (CD79b) homodimer | | Descriptor: | B-cell antigen receptor complex-associated protein beta chain, SULFATE ION | | Authors: | Radaev, S, Sun, P.D. | | Deposit date: | 2009-10-30 | | Release date: | 2010-08-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural and Functional Studies of Igalphabeta and Its Assembly with the B Cell Antigen Receptor.
Structure, 18, 2010
|
|
1WZ9
 
 | | The 2.1 A structure of a tumour suppressing serpin | | Descriptor: | Maspin precursor, SULFATE ION | | Authors: | Law, R.H, Irving, J.A, Buckle, A.M, Ruzyla, K, Buzza, M, Bashtannyk-Puhalovich, T.A, Beddoe, T.C, Kim, N, Worrall, D.M, Bottomley, S.P, Bird, P.I, Rossjohn, J, Whisstock, J.C. | | Deposit date: | 2005-03-03 | | Release date: | 2005-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The high resolution crystal structure of the human tumor suppressor maspin reveals a novel conformational switch in the G-helix.
J.Biol.Chem., 280, 2005
|
|
1XU8
 
 | | The 2.8 A structure of a tumour suppressing serpin | | Descriptor: | Maspin, SULFATE ION | | Authors: | Irving, J.A, Law, R.H, Ruzyla, K, Bashtannyk-Puhalovich, T.A, Kim, N, Worrall, D.M, Rossjohn, J, Whisstock, J.C. | | Deposit date: | 2004-10-25 | | Release date: | 2005-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The high resolution crystal structure of the human tumor suppressor maspin reveals a novel conformational switch in the G-helix.
J.Biol.Chem., 280, 2005
|
|
5AG8
 
 | | CRYSTAL STRUCTURE OF A MUTANT (665I6H) OF THE C-TERMINAL DOMAIN OF RGPB | | Descriptor: | GINGIPAIN R2, GLYCEROL, SULFATE ION | | Authors: | de Diego, I, Ksiazek, M, Mizgalska, D, Golik, P, Szmigielski, B, Nowak, M, Nowakowska, Z, Potempa, B, Koneru, L, Nguyen, K.A, Enghild, J, Thogersen, I.B, Dubin, G, Gomis-Ruth, F.X, Potempa, J. | | Deposit date: | 2015-01-29 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Outer-Membrane Export Signal of Porphyromonas Gingivalis Type Ix Secretion System (T9Ss) is a Conserved C-Terminal Beta-Sandwich Domain.
Sci.Rep., 6, 2016
|
|
6PU9
 
 | | Crystal Structure of the Type B Chloramphenicol O-Acetyltransferase from Vibrio vulnificus | | Descriptor: | 1,2-ETHANEDIOL, Acetyltransferase, CHLORIDE ION | | Authors: | Kim, Y, Maltseva, N, Mulligan, R, Grimshaw, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-07-17 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional characterization of three Type B and C chloramphenicol acetyltransferases from Vibrio species.
Protein Sci., 29, 2020
|
|
6PUA
 
 | | The 2.0 A Crystal Structure of the Type B Chloramphenicol Acetyltransferase from Vibrio cholerae | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Kim, Y, Maltseva, N, Stam, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-07-18 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional characterization of three Type B and C chloramphenicol acetyltransferases from Vibrio species.
Protein Sci., 29, 2020
|
|
4EJN
 
 | | Crystal structure of autoinhibited form of AKT1 in complex with N-(4-(5-(3-acetamidophenyl)-2-(2-aminopyridin-3-yl)-3H-imidazo[4,5-b]pyridin-3-yl)benzyl)-3-fluorobenzamide | | Descriptor: | 1,2-ETHANEDIOL, 2-BUTANOL, N-(4-{5-[3-(acetylamino)phenyl]-2-(2-aminopyridin-3-yl)-3H-imidazo[4,5-b]pyridin-3-yl}benzyl)-3-fluorobenzamide, ... | | Authors: | Eathiraj, S. | | Deposit date: | 2012-04-06 | | Release date: | 2012-05-23 | | Last modified: | 2013-01-02 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Discovery and optimization of a series of 3-(3-phenyl-3H-imidazo[4,5-b]pyridin-2-yl)pyridin-2-amines: orally bioavailable, selective, and potent ATP-independent Akt inhibitors.
J.Med.Chem., 55, 2012
|
|
5N8C
 
 | | Crystal structure of Pseudomonas aeruginosa LpxC complexed with inhibitor | | Descriptor: | (2~{S})-3-azanyl-2-[[(1~{R})-5-[2-[4-[[2-(hydroxymethyl)imidazol-1-yl]methyl]phenyl]ethynyl]-2,3-dihydro-1~{H}-inden-1-yl]amino]-3-methyl-~{N}-oxidanyl-butanamide, CHLORIDE ION, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Cross, J.B, Ryan, M.D, Zhang, J, Cheng, R.K, Wood, M, Andersen, O.A, Brooks, M, Kwong, J, Barker, J. | | Deposit date: | 2017-02-23 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based discovery of LpxC inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5CTX
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a fragment | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-phenyl-3-[2-(pyridin-3-yl)-1,3-thiazol-5-yl]-2,7-dihydro-6H-pyrazolo[3,4-b]pyridin-6-one, DNA gyrase subunit B, ... | | Authors: | Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C, Mesleh, M, Cross, J.B, Zhang, J, Yang, Q, Lippa, B, Ryan, M.D. | | Deposit date: | 2015-07-24 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fragment-based discovery of DNA gyrase inhibitors targeting the ATPase subunit of GyrB.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
