8D2T
 
 | | Zebrafish MFSD2A isoform B in inward open ligand-free conformation | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FAB heavy chain, FAB light chain, ... | | Authors: | Nguyen, C, Lei, H.T, Lai, L.T.F, Gallentino, M.J, Mu, X, Matthies, D, Gonen, T. | | Deposit date: | 2022-05-30 | | Release date: | 2023-05-10 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Lipid flipping in the omega-3 fatty-acid transporter.
Nat Commun, 14, 2023
|
|
8D2V
 
 | | Zebrafish MFSD2A isoform B in inward open ligand 1B conformation | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FAB heavy chain, FAB light chain, ... | | Authors: | Nguyen, C, Lei, H.T, Lai, L.T.F, Gallentino, M.J, Mu, X, Matthies, D, Gonen, T. | | Deposit date: | 2022-05-30 | | Release date: | 2023-05-10 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Lipid flipping in the omega-3 fatty-acid transporter.
Nat Commun, 14, 2023
|
|
8D2W
 
 | | Zebrafish MFSD2A isoform B in inward open ligand 2B conformation | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FAB heavy chain, FAB light chain, ... | | Authors: | Nguyen, C, Lei, H.T, Lai, L.T.F, Gallentino, M.J, Mu, X, Matthies, D, Gonen, T. | | Deposit date: | 2022-05-30 | | Release date: | 2023-05-10 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Lipid flipping in the omega-3 fatty-acid transporter.
Nat Commun, 14, 2023
|
|
8D2S
 
 | | Zebrafish MFSD2A isoform B in inward open ligand bound conformation | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FAB heavy chain, FAB light chain, ... | | Authors: | Nguyen, C, Lei, H.T, Lai, L.T.F, Gallentino, M.J, Mu, X, Matthies, D, Gonen, T. | | Deposit date: | 2022-05-30 | | Release date: | 2023-05-10 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Lipid flipping in the omega-3 fatty-acid transporter.
Nat Commun, 14, 2023
|
|
8D2X
 
 | | Zebrafish MFSD2A isoform B in inward open ligand 3C conformation | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FAB heavy chain, FAB light chain, ... | | Authors: | Nguyen, C, Lei, H.T, Lai, L.T.F, Gallentino, M.J, Mu, X, Matthies, D, Gonen, T. | | Deposit date: | 2022-05-30 | | Release date: | 2023-05-10 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Lipid flipping in the omega-3 fatty-acid transporter.
Nat Commun, 14, 2023
|
|
8D2U
 
 | | Zebrafish MFSD2A isoform B in inward open ligand 1A conformation | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FAB heavy chain, FAB light chain, ... | | Authors: | Nguyen, C, Lei, H.T, Lai, L.T.F, Gallentino, M.J, Mu, X, Matthies, D, Gonen, T. | | Deposit date: | 2022-05-30 | | Release date: | 2023-05-10 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Lipid flipping in the omega-3 fatty-acid transporter.
Nat Commun, 14, 2023
|
|
4KEH
 
 | | Crosslinked Crystal Structure of Type II Fatty Synthase Dehydratase, FabA, and Acyl Carrier Protein, AcpP | | Descriptor: | Acyl carrier protein, N-{3-[DIHYDROXY(NONYL)-LAMBDA~4~-SULFANYL]PROPYL}-N~3~-[(2R)-2-HYDROXY-3,3-DIMETHYL-4-(PHOSPHONOOXY)BUTANOYL]-BETA-ALANINAMIDE, N-{3-[dihydroxy(nonyl)-lambda~4~-sulfanyl]propyl}-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide | | Authors: | Nguyen, C, Haushalter, R, Finzel, K, Leong, J, Le, B.C, Burkart, M, Tsai, S.C. | | Deposit date: | 2013-04-25 | | Release date: | 2013-12-25 | | Last modified: | 2014-01-29 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Trapping the dynamic acyl carrier protein in fatty acid biosynthesis.
Nature, 505, 2014
|
|
1VYQ
 
 | | Novel inhibitors of Plasmodium Falciparum dUTPase provide a platform for anti-malarial drug design | | Descriptor: | 2,3-DEOXY-3-FLUORO-5-O-TRITYLURIDINE, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE | | Authors: | Whittingham, J.L, Leal, I, Kasinathan, G, Nguyen, C, Bell, E, Jones, A.F, Berry, C, Benito, A, Turkenburg, J.P, Dodson, E.J, Ruiz Perez, L.M, Wilkinson, A.J, Johansson, N.G, Brun, R, Gilbert, I.H, Gonzalez Pacanowska, D, Wilson, K.S. | | Deposit date: | 2004-05-05 | | Release date: | 2005-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dutpase as a Platform for Antimalarial Drug Design: Structural Basis for the Selectivity of a Class of Nucleoside Inhibitors.
Structure, 13, 2005
|
|
3JW1
 
 | | Crystal Structure of Bovine Pancreatic Ribonuclease Complexed with Uridine-5'-monophosphate at 1.60 A Resolution | | Descriptor: | Ribonuclease pancreatic, URIDINE-5'-MONOPHOSPHATE | | Authors: | Larson, S.B, Day, J.S, Nguyen, C, Cudney, R, Mcpherson, A, Center for High-Throughput Structural Biology (CHTSB) | | Deposit date: | 2009-09-17 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of bovine pancreatic ribonuclease complexed with uridine 5'-monophosphate at 1.60 A resolution.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3ODQ
 
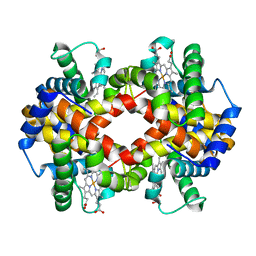 | | Structure of a Crystal Form of Human Methemoglobin Indicative of Fiber Formation | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Larson, S.B, Day, J.S, Nguyen, C, Cudney, R, Mcpherson, A, Center for High-Throughput Structural Biology (CHTSB) | | Deposit date: | 2010-08-11 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of a crystal form of human methemoglobin indicative of fiber formation.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3DYB
 
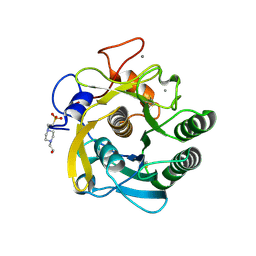 | | proteinase K- digalacturonic acid complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, Proteinase K, ... | | Authors: | Larson, S.B, Day, J.S, McPherson, A, Cudney, R, Nguyen, C, Center for High-Throughput Structural Biology (CHTSB) | | Deposit date: | 2008-07-25 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | High-resolution structure of proteinase K cocrystallized with digalacturonic acid.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3ENJ
 
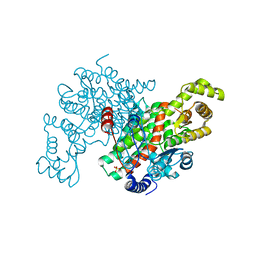 | | Structure of Pig Heart Citrate Synthase at 1.78 A resolution | | Descriptor: | CHLORIDE ION, CYSTEINE, Citrate synthase, ... | | Authors: | Larson, S.B, Day, J.S, Nguyen, C, Cudney, R, McPherson, A, Center for High-Throughput Structural Biology (CHTSB) | | Deposit date: | 2008-09-25 | | Release date: | 2009-02-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of pig heart citrate synthase at 1.78 A resolution.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
6NHL
 
 | | Crystal structure of QueE from Escherichia coli | | Descriptor: | 7-carboxy-7-deazaguanine synthase, DI(HYDROXYETHYL)ETHER, FE (III) ION, ... | | Authors: | Grell, T.A.J, Bell, B.N, Nguyen, C, Dowling, D.P, Drennan, C.L. | | Deposit date: | 2018-12-23 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Crystal structure of AdoMet radical enzyme 7-carboxy-7-deazaguanine synthase from Escherichia coli suggests how modifications near [4Fe-4S] cluster engender flavodoxin specificity.
Protein Sci., 28, 2019
|
|
3PWY
 
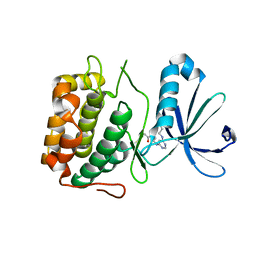 | | Crystal structure of an extender (SPD28345)-modified human PDK1 complex 2 | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, N-[2-({6-[(2-sulfanylethyl)amino]pyrimidin-4-yl}amino)ethyl]propanamide | | Authors: | Elling, R.A, Penny, D.M, Simmons, R.L, Erlanson, D.A, Romanowski, M.J. | | Deposit date: | 2010-12-09 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Discovery of a potent and highly selective PDK1 inhibitor via fragment-based drug discovery.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3QC4
 
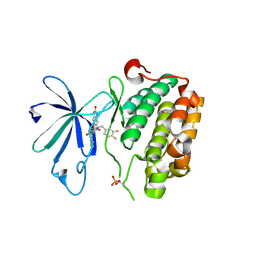 | | PDK1 in complex with DFG-OUT inhibitor xxx | | Descriptor: | 1-(3,4-difluorobenzyl)-2-oxo-N-{(1R)-2-[(2-oxo-2,3-dihydro-1H-benzimidazol-5-yl)oxy]-1-phenylethyl}-1,2-dihydropyridine-3-carboxamide, 3-phosphoinositide-dependent protein kinase 1 | | Authors: | Arndt, J.W. | | Deposit date: | 2011-01-15 | | Release date: | 2011-04-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a potent and highly selective PDK1 inhibitor via fragment-based drug discovery.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
8DNT
 
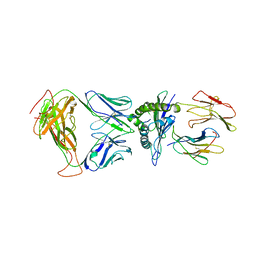 | | SARS-CoV-2 specific T cell receptor | | Descriptor: | Beta-2-microglobulin, MHC class I antigen alpha chain, Nucleoprotein, ... | | Authors: | Gallagher, D.T, Wu, D, Gowthaman, R, Pierce, B.G, Mariuzza, R.A, Weng, N.P. | | Deposit date: | 2022-07-11 | | Release date: | 2023-07-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | SARS-CoV-2 infection establishes a stable and age-independent CD8 + T cell response against a dominant nucleocapsid epitope using restricted T cell receptors.
Nat Commun, 14, 2023
|
|
3B6Z
 
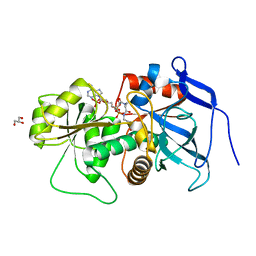 | | Lovastatin polyketide enoyl reductase (LovC) complexed with 2'-phosphoadenosyl isomer of crotonoyl-CoA | | Descriptor: | Enoyl reductase, GLYCEROL, S-{(9R,13R,15S)-17-[(2R,3R,4R,5R)-5-(6-amino-9H-purin-9-yl)-3-hydroxy-4-(phosphonooxy)tetrahydrofuran-2-yl]-9,13,15-trihydroxy-10,10-dimethyl-13,15-dioxido-4,8-dioxo-12,14,16-trioxa-3,7-diaza-13,15-diphosphaheptadec-1-yl}(2E)-but-2-enethioate | | Authors: | Ames, B.D, Smith, P.T, Ma, S.M, Wong, E.W, Xie, X, Vederas, J.C, Tang, Y, Tsai, S.-C. | | Deposit date: | 2007-10-29 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure and biochemical studies of the trans-acting polyketide enoyl reductase LovC from lovastatin biosynthesis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1RII
 
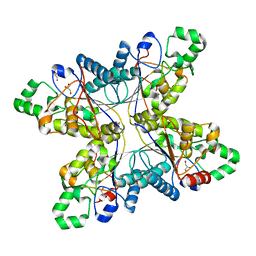 | | Crystal structure of phosphoglycerate mutase from M. Tuberculosis | | Descriptor: | 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase, GLYCEROL | | Authors: | Mueller, P, Sawaya, M.R, Chan, S, Wu, Y, Pashkova, I, Perry, J, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-11-17 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The 1.70 angstroms X-ray crystal structure of Mycobacterium tuberculosis phosphoglycerate mutase.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
6P3P
 
 | | Crystal structure of Mcl-1 in complex with compound 65 | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1, methyl N-(5-{[2-chloro-5-(trifluoromethyl)phenyl]sulfamoyl}-4-methylthiophene-2-carbonyl)-D-phenylalaninate | | Authors: | Toms, A.V, Follows, B. | | Deposit date: | 2019-05-24 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Discovery of novel biaryl sulfonamide based Mcl-1 inhibitors.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
5ZE4
 
 | | The structure of holo- structure of DHAD complex with [2Fe-2S] cluster | | Descriptor: | ACETATE ION, Dihydroxy-acid dehydratase, chloroplastic, ... | | Authors: | Zhou, J, Zang, X, Tang, Y, Yan, Y, Gan, J, Wu, L. | | Deposit date: | 2018-02-26 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Resistance-gene-directed discovery of a natural-product herbicide with a new mode of action.
Nature, 559, 2018
|
|
3B70
 
 | | Crystal structure of Aspergillus terreus trans-acting lovastatin polyketide enoyl reductase (LovC) with bound NADP | | Descriptor: | Enoyl reductase, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ames, B.D, Smith, P.T, Ma, S.M, Wong, E.W, Xie, X, Vederas, J.C, Tang, Y, Tsai, S.-C. | | Deposit date: | 2007-10-29 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure and biochemical studies of the trans-acting polyketide enoyl reductase LovC from lovastatin biosynthesis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
