4WD7
 
 | |
5IWS
 
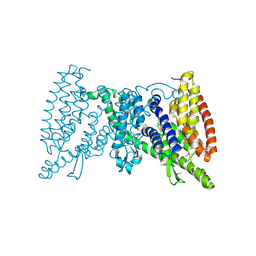 | | Crystal structure of the transporter MalT, the EIIC domain from the maltose-specific phosphotransferase system | | Descriptor: | Protein-N(Pi)-phosphohistidine-sugar phosphotransferase (Enzyme II of the phosphotransferase system) (PTS system glucose-specific IIBC component), alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | McCoy, J.G, Ren, Z, Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2016-03-22 | | Release date: | 2016-05-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | The Structure of a Sugar Transporter of the Glucose EIIC Superfamily Provides Insight into the Elevator Mechanism of Membrane Transport.
Structure, 24, 2016
|
|
4RYQ
 
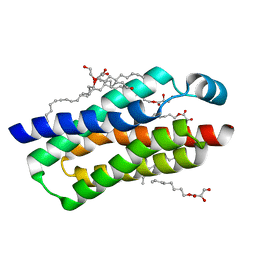 | | Crystal structure of BcTSPO, type 2 at 1.7 Angstrom | | Descriptor: | Integral membrane protein, [(Z)-octadec-9-enyl] (2R)-2,3-bis(oxidanyl)propanoate | | Authors: | Guo, Y, Liu, Q, Hendrickson, W.A, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-12-16 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Protein structure. Structure and activity of tryptophan-rich TSPO proteins.
Science, 347, 2015
|
|
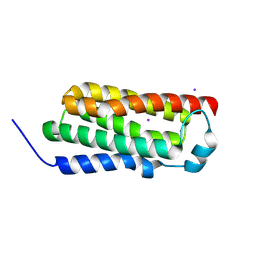
4RYM
 
 | |
4RYO
 
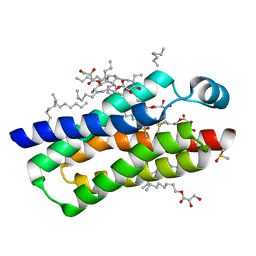 | | Crystal structure of BcTSPO type II high resolution monomer | | Descriptor: | DIMETHYL SULFOXIDE, Integral membrane protein, [(Z)-octadec-9-enyl] (2R)-2,3-bis(oxidanyl)propanoate | | Authors: | Guo, Y, Liu, Q, Hendrickson, W.A, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-12-15 | | Release date: | 2015-04-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Protein structure. Structure and activity of tryptophan-rich TSPO proteins.
Science, 347, 2015
|
|
4RYR
 
 | | Crystal structure of BcTSPO, type 2 at 1.7 Angstrom with DMSO | | Descriptor: | DIMETHYL SULFOXIDE, Integral membrane protein, [(Z)-octadec-9-enyl] (2R)-2,3-bis(oxidanyl)propanoate | | Authors: | Guo, Y, Liu, Q, Hendrickson, W.A, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-12-16 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | Protein structure. Structure and activity of tryptophan-rich TSPO proteins.
Science, 347, 2015
|
|
4RYN
 
 | | Crystal structure of BcTSPO, type1 monomer | | Descriptor: | CACODYLATE ION, DODECYL-ALPHA-D-MALTOSIDE, Integral membrane protein, ... | | Authors: | Guo, Y, Liu, Q, Hendrickson, W.A, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-12-15 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Protein structure. Structure and activity of tryptophan-rich TSPO proteins.
Science, 347, 2015
|
|
4RYJ
 
 | |
4RYI
 
 | |
4TQ6
 
 | | Structure of a UbiA homolog from Archaeoglobus fulgidus bound to Cd2+ | | Descriptor: | CADMIUM ION, prenyltransferase | | Authors: | Huang, H, Levin, E.J, Bai, Y, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-06-10 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.0678 Å) | | Cite: | Structure of a Membrane-Embedded Prenyltransferase Homologous to UBIAD1.
Plos Biol., 12, 2014
|
|
4TQ5
 
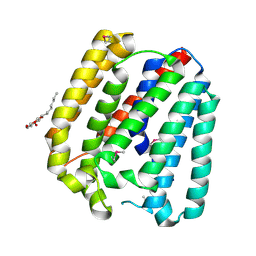 | | Structure of a UbiA homolog from Archaeoglobus fulgidus | | Descriptor: | octyl beta-D-glucopyranoside, prenyltransferase | | Authors: | Huang, H, Levin, E.J, Bai, Y, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-06-10 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2023 Å) | | Cite: | Structure of a Membrane-Embedded Prenyltransferase Homologous to UBIAD1.
Plos Biol., 12, 2014
|
|
4TQ4
 
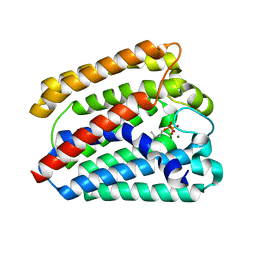 | | Structure of a UbiA homolog from Archaeoglobus fulgidus bound to DMAPP and Mg2+ | | Descriptor: | DIMETHYLALLYL DIPHOSPHATE, MAGNESIUM ION, prenyltransferase | | Authors: | Huang, H, Levin, E.J, Bai, Y, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-06-10 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5025 Å) | | Cite: | Structure of a Membrane-Embedded Prenyltransferase Homologous to UBIAD1.
Plos Biol., 12, 2014
|
|
4TQ3
 
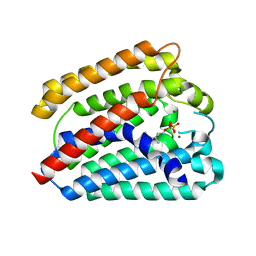 | | Structure of a UbiA homolog from Archaeoglobus fulgidus bound to GPP and Mg2+ | | Descriptor: | GERANYL DIPHOSPHATE, MAGNESIUM ION, Prenyltransferase | | Authors: | Huang, H, Levin, E.J, Bai, Y, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-06-10 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4076 Å) | | Cite: | Structure of a Membrane-Embedded Prenyltransferase Homologous to UBIAD1.
Plos Biol., 12, 2014
|
|
5F15
 
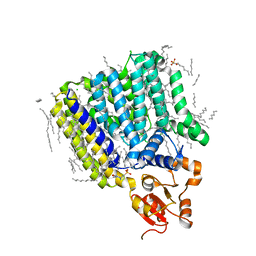 | | Crystal Structure of ArnT from Cupriavidus metallidurans bound to Undecaprenyl phosphate | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-amino-4-deoxy-L-arabinose (L-Ara4N) transferase, CHLORIDE ION, ... | | Authors: | Petrou, V.I, Clarke, O.B, Tomasek, D, Banerjee, S, Rajashankar, K.R, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2015-11-30 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of aminoarabinose transferase ArnT suggest a molecular basis for lipid A glycosylation.
Science, 351, 2016
|
|
5EZM
 
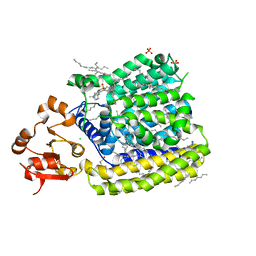 | | Crystal Structure of ArnT from Cupriavidus metallidurans in the apo state | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferases of PMT family, CHLORIDE ION, ... | | Authors: | Petrou, V.I, Clarke, O.B, Tomasek, D, Banerjee, S, Rajashankar, K.R, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2015-11-26 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of aminoarabinose transferase ArnT suggest a molecular basis for lipid A glycosylation.
Science, 351, 2016
|
|
6WTW
 
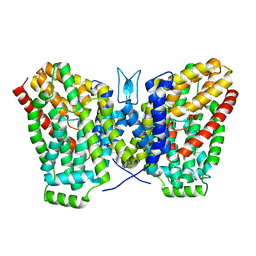 | | Structure of LaINDY crystallized in the presence of alpha-ketoglutarate and malate | | Descriptor: | DASS family sodium-coupled anion symporter | | Authors: | Sauer, D.B, Cocco, N, Marden, J.J, Song, J.M, Wang, D.N, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2020-05-04 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural basis for the reaction cycle of DASS dicarboxylate transporters.
Elife, 9, 2020
|
|
4TKR
 
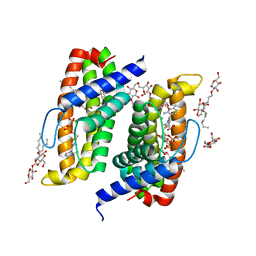 | | Native-SAD phasing for ThiT from Listeria monocytogenes serovar. | | Descriptor: | 2-O-octyl-beta-D-glucopyranose, THIAMINE DIPHOSPHATE, Thiamine transporter thia | | Authors: | Guo, Y, Liu, Q, Hendrickson, W.A, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-05-27 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.0023 Å) | | Cite: | Multi-crystal native SAD analysis at 6 keV.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4TKQ
 
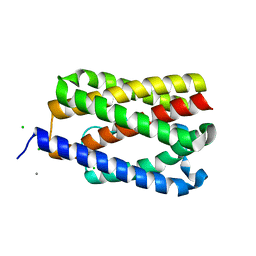 | | Native-SAD phasing for YetJ from Bacillus Subtilis | | Descriptor: | CALCIUM ION, CHLORIDE ION, Uncharacterized protein YetJ | | Authors: | Liu, Q, Chang, Y, Hendrickson, W.A, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2014-05-27 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8025 Å) | | Cite: | Multi-crystal native SAD analysis at 6 keV.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4WD8
 
 | |
5EKE
 
 | | Structure of the polyisoprenyl-phosphate glycosyltransferase GtrB (F215A mutant) | | Descriptor: | MAGNESIUM ION, URIDINE-5'-DIPHOSPHATE, Uncharacterized glycosyltransferase sll0501 | | Authors: | Ardiccioni, C, Clarke, O.B, Tomasek, D, Banerjee, S, Rajashankar, K.R, Liu, Q, Shapiro, L, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2015-11-03 | | Release date: | 2016-01-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Structure of the polyisoprenyl-phosphate glycosyltransferase GtrB and insights into the mechanism of catalysis.
Nat Commun, 7, 2016
|
|
6WU1
 
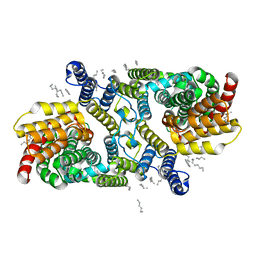 | | Structure of apo LaINDY | | Descriptor: | DASS family sodium-coupled anion symporter, DECANE, HEXANE, ... | | Authors: | Sauer, D.B, Marden, J.J, Cocco, N.C, Song, J.M, Wang, D.N, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2020-05-04 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis for the reaction cycle of DASS dicarboxylate transporters.
Elife, 9, 2020
|
|
6WU2
 
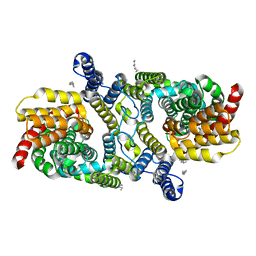 | | Structure of the LaINDY-malate complex | | Descriptor: | DASS family sodium-coupled anion symporter, DECANE, HEXANE, ... | | Authors: | Sauer, D.B, Marden, J.J, Cocco, N, Song, J.M, Wang, D.N, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2020-05-04 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural basis for the reaction cycle of DASS dicarboxylate transporters.
Elife, 9, 2020
|
|
6WU4
 
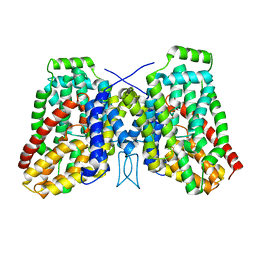 | | Structure of the LaINDY-alpha-ketoglutarate complex | | Descriptor: | DASS family sodium-coupled anion symporter | | Authors: | Sauer, D.B, Marden, J.J, Cocco, N, Song, J.M, Wang, D.N, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2020-05-04 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural basis for the reaction cycle of DASS dicarboxylate transporters.
Elife, 9, 2020
|
|
6WM5
 
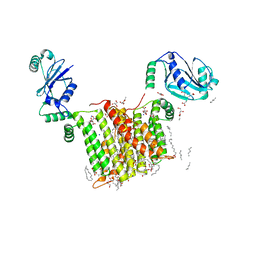 | | Structure of a phosphatidylinositol-phosphate synthase (PIPS) from Mycobacterium kansasii | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, 3,3',3''-phosphanetriyltripropanoic acid, ... | | Authors: | Belcher Dufrisne, M, Jorge, C.D, Timoteo, C.G, Petrou, V.I, Ashraf, K.U, Banerjee, S, Clarke, O.B, Santos, H, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2020-04-20 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.961 Å) | | Cite: | Structural and Functional Characterization of Phosphatidylinositol-Phosphate Biosynthesis in Mycobacteria.
J.Mol.Biol., 432, 2020
|
|
2K2B
 
 | |
