3DIF
 
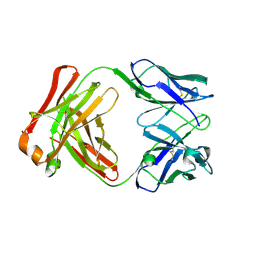 | |
4I2X
 
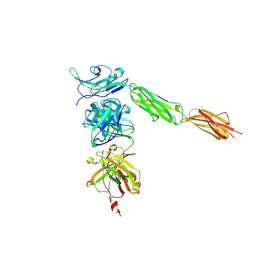 | | Crystal structure of Signal Regulatory Protein gamma (SIRP-gamma) in complex with FabOX117 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, FabOX117 heavy chain, ... | | Authors: | Nettleship, J.E, Ren, J, Stuart, D.I, Owens, R.J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2012-11-23 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structure of signal regulatory protein gamma (SIRP gamma) in complex with an antibody Fab fragment.
Bmc Struct.Biol., 13, 2013
|
|
3DGG
 
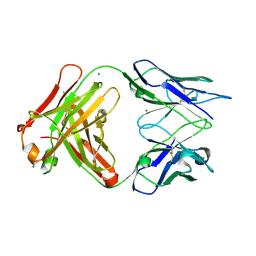 | | Crystal structure of FabOX108 | | Descriptor: | FabOX108 Heavy Chain Fragment, FabOX108 Light Chain Fragment, MAGNESIUM ION | | Authors: | Ren, J, Nettleship, J.E, Owens, R.J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2008-06-13 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A pipeline for the production of antibody fragments for structural studies using transient expression in HEK 293T cells.
Protein Expr.Purif., 62, 2008
|
|
4KQQ
 
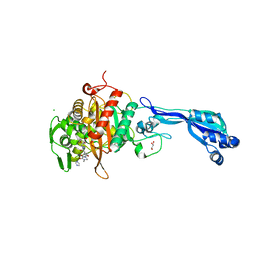 | | CRYSTAL STRUCTURE OF PENICILLIN-BINDING PROTEIN 3 FROM PSEUDOMONAS AERUGINOSA IN COMPLEX WITH (5S)-Penicilloic Acid | | Descriptor: | (2S,4S)-2-[(R)-carboxy{[(2R)-2-{[(4-ethyl-2,3-dioxopiperazin-1-yl)carbonyl]amino}-2-phenylacetyl]amino}methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nettleship, J.E, Stuart, D.I, Owens, R.J, Ren, J. | | Deposit date: | 2013-05-15 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding of (5S)-Penicilloic Acid to Penicillin Binding Protein 3.
Acs Chem.Biol., 8, 2013
|
|
4KQO
 
 | | Crystal structure of penicillin-binding protein 3 from pseudomonas aeruginosa in complex with piperacillin | | Descriptor: | CHLORIDE ION, GLYCEROL, IMIDAZOLE, ... | | Authors: | Nettleship, J.E, Stuart, D.I, Owens, R.J, Ren, J. | | Deposit date: | 2013-05-15 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Binding of (5S)-Penicilloic Acid to Penicillin Binding Protein 3.
Acs Chem.Biol., 8, 2013
|
|
4KQR
 
 | | CRYSTAL STRUCTURE OF PENICILLIN-BINDING PROTEIN 3 FROM PSEUDOMONAS AERUGINOSA IN COMPLEX WITH (5S)-Penicilloic Acid | | Descriptor: | (2S,4S)-2-[(R)-carboxy{[(2R)-2-{[(4-ethyl-2,3-dioxopiperazin-1-yl)carbonyl]amino}-2-phenylacetyl]amino}methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nettleship, J.E, Stuart, D.I, Owens, R.J, Ren, J. | | Deposit date: | 2013-05-15 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Binding of (5S)-Penicilloic Acid to Penicillin Binding Protein 3.
Acs Chem.Biol., 8, 2013
|
|
4UXJ
 
 | | Leishmania major Thymidine Kinase in complex with dTTP | | Descriptor: | MAGNESIUM ION, THYMIDINE KINASE, THYMIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Timm, J, Bosch-Navarrete, C, Recio, E, Nettleship, J.E, Rada, H, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2014-08-22 | | Release date: | 2015-05-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Kinetic Characterization of Thymidine Kinase from Leishmania Major.
Plos Negl Trop Dis, 9, 2015
|
|
2A0J
 
 | | Crystal Structure of Nitrogen Regulatory Protein IIA-Ntr from Neisseria meningitidis | | Descriptor: | PTS system, nitrogen regulatory IIA protein | | Authors: | Ren, J, Sainsbury, S, Berrow, N.S, Alderton, D, Nettleship, J.E, Stammers, D.K, Saunders, N.J, Owens, R.J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2005-06-16 | | Release date: | 2005-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of nitrogen regulatory protein IIANtr from Neisseria meningitidis
Bmc Struct.Biol., 5, 2005
|
|
4UXH
 
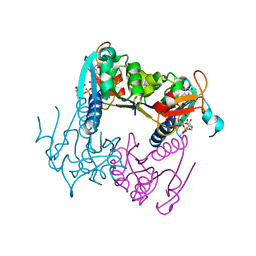 | | Leishmania major Thymidine Kinase in complex with AP5dT | | Descriptor: | P1-(5'-ADENOSYL)P5-(5'-THYMIDYL)PENTAPHOSPHATE, THYMIDINE KINASE, ZINC ION | | Authors: | Timm, J, Bosch-Navarrete, C, Recio, E, Nettleship, J.E, Rada, H, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2014-08-22 | | Release date: | 2015-05-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Kinetic Characterization of Thymidine Kinase from Leishmania Major.
Plos Negl Trop Dis, 9, 2015
|
|
4UXI
 
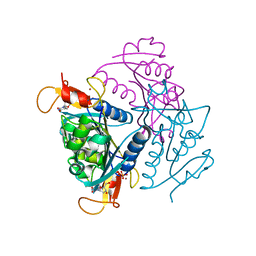 | | Leishmania major Thymidine Kinase in complex with thymidine | | Descriptor: | PHOSPHATE ION, THYMIDINE, THYMIDINE KINASE, ... | | Authors: | Timm, J, Bosch-Navarrete, C, Recio, E, Nettleship, J.E, Rada, H, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2014-08-22 | | Release date: | 2015-05-27 | | Last modified: | 2016-03-02 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural and Kinetic Characterization of Thymidine Kinase from Leishmania Major.
Plos Negl Trop Dis, 9, 2015
|
|
5ML9
 
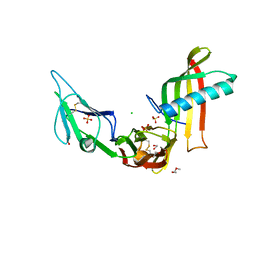 | | Cocrystal structure of Fc gamma receptor IIIa interacting with Affimer F4, a specific binding protein which blocks IgG binding to the receptor. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Affimer F4 with specificity for Fc gamma receptor IIIa, CHLORIDE ION, ... | | Authors: | Robinson, J.I, Tomlinson, D.C, Baxter, E.W, Owen, R.L, Thomsen, M, Win, S.J, Nettleship, J.E, Tiede, C, Foster, R.J, Waterhouse, M.P, Harris, S.A, Owens, R.J, Fishwick, C.W.G, Goldman, A, McPherson, M.J, Morgan, A.W. | | Deposit date: | 2016-12-06 | | Release date: | 2017-12-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Affimer proteins inhibit immune complex binding to Fc gamma RIIIa with high specificity through competitive and allosteric modes of action.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5MN2
 
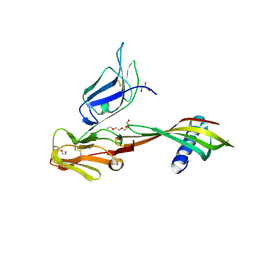 | | Cocrystal structure of Fc gamma receptor IIIa interacting with Affimer G3, a specific binding protein which blocks IgG binding to the receptor. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Affimer G3, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Robinson, J.I, Owen, R.L, Tomlinson, D.C, Baxter, E.W, Nettleship, J.E, Waterhouse, M.P, Harris, S.A, Owens, R.J, McPherson, M.J, Morgan, A.W, Tiede, C, Goldman, A, Thomsen, M. | | Deposit date: | 2016-12-12 | | Release date: | 2017-12-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Affimer proteins inhibit immune complex binding to Fc gamma RIIIa with high specificity through competitive and allosteric modes of action.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3KXA
 
 | | Crystal Structure of NGO0477 from Neisseria gonorrhoeae | | Descriptor: | ASPARAGINE, CHLORIDE ION, Putative uncharacterized protein, ... | | Authors: | Ren, J, Sainsbury, S, Nettleship, J.E, Owens, R.J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2009-12-02 | | Release date: | 2010-01-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of NGO0477 from Neisseria gonorrhoeae reveals a novel protein fold incorporating a helix-turn-helix motif.
Proteins, 78, 2010
|
|
7OFZ
 
 | | Nontypeable Haemophillus influenzae SapA in complex with double stranded RNA | | Descriptor: | ABC-type transport system, periplasmic component, involved in antimicrobial peptide resistance, ... | | Authors: | Lukacik, P, Owen, C.D, Nettleship, J.E, Bird, L.E, Owens, R.J, Walsh, M.A. | | Deposit date: | 2021-05-05 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | The structure of nontypeable Haemophilus influenzae SapA in a closed conformation reveals a constricted ligand-binding cavity and a novel RNA binding motif.
Plos One, 16, 2021
|
|
7OG0
 
 | | Nontypeable Haemophillus influenzae SapA in open and closed conformations, in complex with double stranded RNA | | Descriptor: | ABC-type transport system, periplasmic component, involved in antimicrobial peptide resistance, ... | | Authors: | Lukacik, P, Owen, C.D, Nettleship, J.E, Bird, L.E, Owens, R.J, Walsh, M.A. | | Deposit date: | 2021-05-05 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | The structure of nontypeable Haemophilus influenzae SapA in a closed conformation reveals a constricted ligand-binding cavity and a novel RNA binding motif.
Plos One, 16, 2021
|
|
7OFW
 
 | | Nontypeable Haemophillus influenzae SapA in complex with heme | | Descriptor: | ABC-type transport system, periplasmic component, involved in antimicrobial peptide resistance, ... | | Authors: | Lukacik, P, Owen, C.D, Nettleship, J.E, Bird, L.E, Owens, R.J, Walsh, M.A. | | Deposit date: | 2021-05-05 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The structure of nontypeable Haemophilus influenzae SapA in a closed conformation reveals a constricted ligand-binding cavity and a novel RNA binding motif.
Plos One, 16, 2021
|
|
6QNA
 
 | | Structure of bovine anti-RSV hybrid Fab B13HC-B4LC | | Descriptor: | B13 Heavy chain, B4 light chain, GLYCEROL | | Authors: | Ren, J, Nettleship, J.E, Harris, G, Mwangi, W, Rhaman, N, Grant, C, Kotecha, A, Fry, E, Charleston, B, Stuart, D.I, Hammond, J, Owens, R.J. | | Deposit date: | 2019-02-10 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | The role of the light chain in the structure and binding activity of two cattle antibodies that neutralize bovine respiratory syncytial virus.
Mol.Immunol., 112, 2019
|
|
6QN9
 
 | | Structure of bovine anti-RSV Fab B4 | | Descriptor: | GLYCEROL, Heavy chain, SULFATE ION, ... | | Authors: | Ren, J, Nettleship, J.E, Harris, G, Mwangi, W, Rhaman, N, Grant, C, Kotecha, A, Fry, E, Charleston, B, Stuart, D.I, Hammond, J, Owens, R.J. | | Deposit date: | 2019-02-10 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The role of the light chain in the structure and binding activity of two cattle antibodies that neutralize bovine respiratory syncytial virus.
Mol.Immunol., 112, 2019
|
|
6QN7
 
 | | Structure of bovine anti-RSV hybrid Fab B4HC-B13LC | | Descriptor: | Heavy chain of bovine anti-RSV B4, Light chain of bovine anti-RSV B13 | | Authors: | Ren, J, Nettleship, J.E, Harris, G, Mwangi, W, Rhaman, N, Grant, C, Kotecha, A, Fry, E, Charleston, B, Stuart, D.I, Hammond, J, Owens, R.J. | | Deposit date: | 2019-02-10 | | Release date: | 2019-05-29 | | Last modified: | 2019-08-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The role of the light chain in the structure and binding activity of two cattle antibodies that neutralize bovine respiratory syncytial virus.
Mol.Immunol., 112, 2019
|
|
6QN8
 
 | | Structure of bovine anti-RSV Fab B13 | | Descriptor: | CHLORIDE ION, Heavy chain of bovine anti-RSV B13 Fab, Light chain of bovine anti-RSV Fab B13 | | Authors: | Ren, J, Nettleship, J.E, Harris, G, Mwangi, W, Rhaman, N, Grant, C, Kotecha, A, Fry, E, Charleston, B, Stuart, D.I, Hammond, J, Owens, R.J. | | Deposit date: | 2019-02-10 | | Release date: | 2019-05-29 | | Last modified: | 2019-08-21 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The role of the light chain in the structure and binding activity of two cattle antibodies that neutralize bovine respiratory syncytial virus.
Mol.Immunol., 112, 2019
|
|
4N6U
 
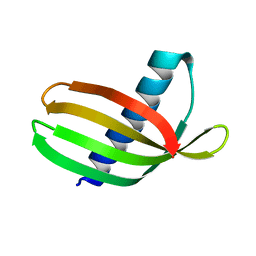 | | Adhiron: a stable and versatile peptide display scaffold - truncated adhiron | | Descriptor: | Adhiron | | Authors: | Mcpherson, M, Tomlinson, D, Owen, R.L, Nettleship, J.E, Owens, R.J. | | Deposit date: | 2013-10-14 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Adhiron: a stable and versatile peptide display scaffold for molecular recognition applications.
Protein Eng.Des.Sel., 27, 2014
|
|
4N6T
 
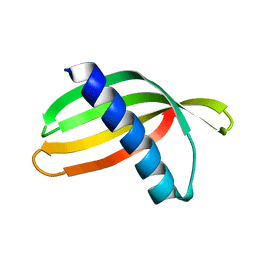 | | Adhiron: a stable and versatile peptide display scaffold - full length adhiron | | Descriptor: | Adhiron | | Authors: | Mcpherson, M, Tomlinson, D, Owen, R.L, Nettleship, J.E, Owens, R.J. | | Deposit date: | 2013-10-14 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Adhiron: a stable and versatile peptide display scaffold for molecular recognition applications.
Protein Eng.Des.Sel., 27, 2014
|
|
2UVD
 
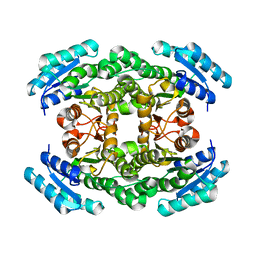 | | The crystal structure of a 3-oxoacyl-(acyl carrier protein) reductase from Bacillus anthracis (BA3989) | | Descriptor: | 3-OXOACYL-(ACYL-CARRIER-PROTEIN) REDUCTASE | | Authors: | Zaccai, N.R, Carter, L.G, Berrow, N.S, Sainsbury, S, Nettleship, J.E, Walter, T.S, Harlos, K, Owens, R.J, Wilson, K.S, Stuart, D.I, Esnouf, R.M, Oxford Protein Production Facility (OPPF), Structural Proteomics in Europe (SPINE) | | Deposit date: | 2007-03-09 | | Release date: | 2007-04-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of a 3-Oxoacyl-(Acylcarrier Protein) Reductase (Ba3989) from Bacillus Anthracis at 2.4-A Resolution.
Proteins: Struct., Funct., Bioinf., 70, 2008
|
|
2VD8
 
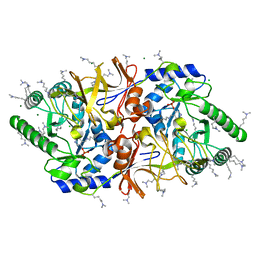 | | The crystal structure of alanine racemase from Bacillus anthracis (BA0252) | | Descriptor: | ALANINE RACEMASE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Au, K, Ren, J, Walter, T.S, Harlos, K, Nettleship, J.E, Owens, R.J, Stuart, D.I, Esnouf, R.M, Oxford Protein Production Facility (OPPF), Structural Proteomics in Europe (SPINE) | | Deposit date: | 2007-10-01 | | Release date: | 2008-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structures of an Alanine Racemase from Bacillus Anthracis (Ba0252) in the Presence and Absence of (R)-1-Aminoethylphosphonic Acid (L-Ala-P).
Acta Crystallogr.,Sect.F, 64, 2008
|
|
2VD9
 
 | | The crystal structure of alanine racemase from Bacillus anthracis (BA0252) with bound L-Ala-P | | Descriptor: | (1S)-1-[((1E)-{3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYLENE)AMINO]ETHYLPHOSPHONIC ACID, ALANINE RACEMASE, CHLORIDE ION, ... | | Authors: | Au, K, Ren, J, Walter, T.S, Harlos, K, Nettleship, J.E, Owens, R.J, Stuart, D.I, Esnouf, R.M, Oxford Protein Production Facility (OPPF), Structural Proteomics in Europe (SPINE) | | Deposit date: | 2007-10-01 | | Release date: | 2008-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of an Alanine Racemase from Bacillus Anthracis (Ba0252) in the Presence and Absence of (R)-1-Aminoethylphosphonic Acid (L-Ala-P).
Acta Crystallogr.,Sect.F, 64, 2008
|
|
