6E5C
 
 | | Solution NMR structure of a de novo designed double-stranded beta-helix | | Descriptor: | De novo beta protein | | Authors: | Marcos, E, Chidyausiku, T.M, McShan, A, Evangelidis, T, Nerli, S, Sgourakis, N, Tripsianes, K, Baker, D. | | Deposit date: | 2018-07-19 | | Release date: | 2018-11-07 | | Last modified: | 2018-11-14 | | Method: | SOLUTION NMR | | Cite: | De novo design of a non-local beta-sheet protein with high stability and accuracy.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7RY6
 
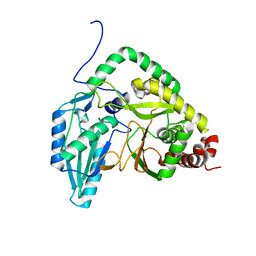 | | Solution NMR structural bundle of the first cyclization domain from yersiniabactin synthetase (Cy1) impacted by dynamics | | Descriptor: | HMWP2 nonribosomal peptide synthetase | | Authors: | Kancherla, A.K, Mishra, S.H, Marincin, K.A, Nerli, S, Sgourakis, N.G, Dowling, D.P, Bouvignies, G, Frueh, D.P. | | Deposit date: | 2021-08-24 | | Release date: | 2022-07-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Global protein dynamics as communication sensors in peptide synthetase domains.
Sci Adv, 8, 2022
|
|
6O0I
 
 | | NMR ensemble of computationally designed protein XAA | | Descriptor: | Design construct XAA | | Authors: | Wei, K.Y, Moschidi, D, Nerli, S, Sgourakis, N, Baker, D. | | Deposit date: | 2019-02-16 | | Release date: | 2020-04-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Computational design of closely related proteins that adopt two well-defined but structurally divergent folds.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6O0C
 
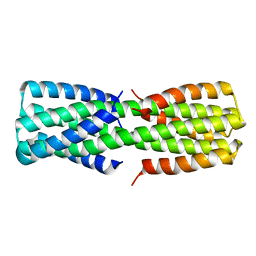 | | NMR ensemble of computationally designed protein XAA_GVDQ mutant M4L | | Descriptor: | Design construct XAA_GVDQ mutant M4L | | Authors: | Wei, K.Y, Moschidi, D, Nerli, S, Sgourakis, N, Baker, D. | | Deposit date: | 2019-02-15 | | Release date: | 2020-04-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Computational design of closely related proteins that adopt two well-defined but structurally divergent folds.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6OSW
 
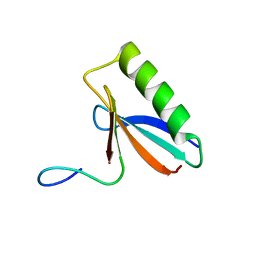 | | An order-to-disorder structural switch activates the FoxM1 transcription factor | | Descriptor: | Forkhead box M1 | | Authors: | Marceau, A.H, Rubin, S.M, Nerli, S, McShane, A.C, Sgourakis, N.G. | | Deposit date: | 2019-05-02 | | Release date: | 2019-05-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | An order-to-disorder structural switch activates the FoxM1 transcription factor.
Elife, 8, 2019
|
|
5WOX
 
 | |
5WOZ
 
 | |
5WOY
 
 | |
5WOT
 
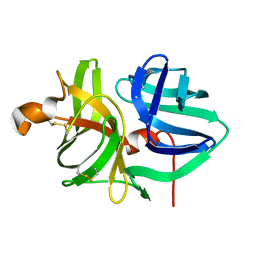 | |
6AT9
 
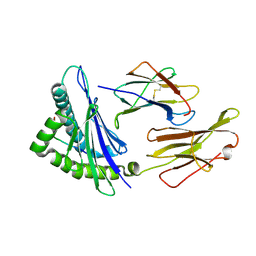 | | Crystal structure of an anaplastic lymphoma kinase-derived neuroblastoma tumor antigen bound to the Human Major Histocompatibility Complex Class I molecule HLA-A*0101 | | Descriptor: | ALK, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Toor, J, Rao, A.A, Salama, S, Tripathi, S, Haussler, D, Sgourakis, N.G. | | Deposit date: | 2017-08-28 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9503 Å) | | Cite: | A Recurrent Mutation in Anaplastic Lymphoma Kinase with Distinct Neoepitope Conformations.
Front Immunol, 9, 2018
|
|
5TXS
 
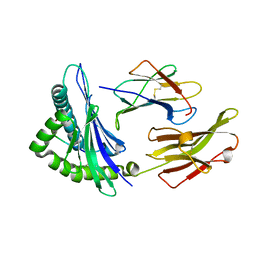 | | Crystal structure of an anaplastic lymphoma kinase-derived neuroblastoma tumor antigen bound to the Human Major Histocompatibility Complex Class I molecule HLA-B*1501 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-15 alpha chain, ... | | Authors: | Toor, J, Rao, A.A, Salama, S, Tripathi, S, Haussler, D, Sgourakis, N.G. | | Deposit date: | 2016-11-17 | | Release date: | 2017-11-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | A Recurrent Mutation in Anaplastic Lymphoma Kinase with Distinct Neoepitope Conformations.
Front Immunol, 9, 2018
|
|
5VZ5
 
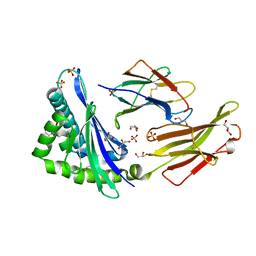 | | Crystal structure of an anaplastic lymphoma kinase-derived neuroblastoma tumor antigen bound to the Human Major Histocompatibility Complex Class I molecule HLA-B*1501 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Toor, J, Rao, A.A, Salama, S, Tripathi, S, Haussler, D, Sgourakis, N.G. | | Deposit date: | 2017-05-26 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5901 Å) | | Cite: | A Recurrent Mutation in Anaplastic Lymphoma Kinase with Distinct Neoepitope Conformations.
Front Immunol, 9, 2018
|
|
7SKO
 
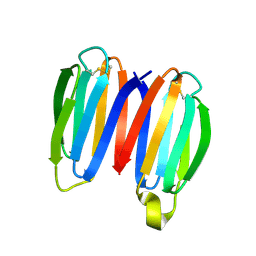 | | De novo synthetic protein DIG8-CC (orthogonal space group) | | Descriptor: | De novo synthetic protein DIG8-CC, MAGNESIUM ION | | Authors: | Mendes, S.R, Eckhard, U, Marcos, E, Gomis-Ruth, F.X. | | Deposit date: | 2021-10-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | De novo design of immunoglobulin-like domains
Nat Commun, 13, 2022
|
|
7SKN
 
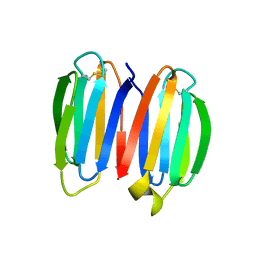 | |
7SKP
 
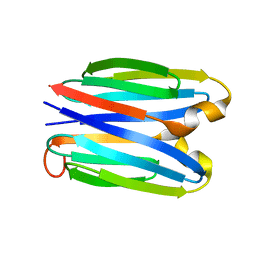 | |
6NY8
 
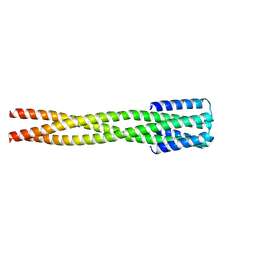 | |
6NYE
 
 | |
6NZ1
 
 | |
6NXM
 
 | |
6NZ3
 
 | |
6NX2
 
 | | Crystal structure of computationally designed protein AAA | | Descriptor: | BROMIDE ION, Design construct AAA | | Authors: | Wei, K.Y, Bick, M.J. | | Deposit date: | 2019-02-07 | | Release date: | 2020-04-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Computational design of closely related proteins that adopt two well-defined but structurally divergent folds.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6NYI
 
 | |
6NYK
 
 | |
