3FG1
 
 | | Crystal structure of Delta413-417:GS LOX | | Descriptor: | ACETIC ACID, Allene oxide synthase-lipoxygenase protein, CALCIUM ION, ... | | Authors: | Neau, D.B, Newcomer, M.E. | | Deposit date: | 2008-12-04 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The 1.85 A structure of an 8R-lipoxygenase suggests a general model for lipoxygenase product specificity.
Biochemistry, 48, 2009
|
|
3FG4
 
 | | Crystal structure of Delta413-417:GS I805A LOX | | Descriptor: | ACETIC ACID, ARACHIDONIC ACID, Allene oxide synthase-lipoxygenase protein, ... | | Authors: | Neau, D.B, Newcomer, M.E. | | Deposit date: | 2008-12-04 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The 1.85 A structure of an 8R-lipoxygenase suggests a general model for lipoxygenase product specificity.
Biochemistry, 48, 2009
|
|
3FG3
 
 | | Crystal structure of Delta413-417:GS I805W LOX | | Descriptor: | ACETIC ACID, Allene oxide synthase-lipoxygenase protein, CALCIUM ION, ... | | Authors: | Neau, D.B, Newcomer, M.E. | | Deposit date: | 2008-12-04 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.85 A structure of an 8R-lipoxygenase suggests a general model for lipoxygenase product specificity.
Biochemistry, 48, 2009
|
|
2EI3
 
 | | Anaerobic Crystal Structure Analysis of the 1,2-dihydroxynaphthalene dioxygenase from Pseudomonas sp. strain C18 complexes with 2,3-dihydroxybiphenyl | | Descriptor: | 1,2-dihydroxynaphthalene dioxygenase, BIPHENYL-2,3-DIOL, FE (II) ION, ... | | Authors: | Neau, D.B, Kelker, M.S, Colbert, C.L, Bolin, J.T. | | Deposit date: | 2007-03-10 | | Release date: | 2008-02-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural explanation for success and failure in the enzymatic ring-cleavage of 3,4 dihydroxybiphenyl and related PCB metabolites
To be Published
|
|
2EI0
 
 | | Anaerobic Crystal Structure Analysis of 1,2-dihydroxynaphthalene dioxygenase from Pseudomonas sp. strain C18 complexed with 3,4-dihydroxybiphenyl | | Descriptor: | 1,1'-BIPHENYL-3,4-DIOL, 1,2-dihydroxynaphthalene dioxygenase, FE (II) ION, ... | | Authors: | Neau, D.B, Kelker, M.S, Colbert, C.L, Bolin, J.T. | | Deposit date: | 2007-03-10 | | Release date: | 2008-02-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural explanation for success and failure in the enzymatic ring-cleavage of 3,4 dihydroxybiphenyl and related PCB metabolites
To be Published
|
|
2EI2
 
 | | Crystal Structure Analysis of the 1,2-dihydroxynaphthalene dioxygenase from Pseudomonas sp. stain C18 | | Descriptor: | 1,2-dihydroxynaphthalene dioxygenase, FE (II) ION, GLYCEROL, ... | | Authors: | Neau, D.B, Kelker, M.S, Colbert, C.L, Bolin, J.T. | | Deposit date: | 2007-03-10 | | Release date: | 2008-02-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural explanation for success and failure in the enzymatic ring-cleavage of 3,4 dihydroxybiphenyl and related PCB metabolites
To be Published
|
|
2EI1
 
 | | Anaerobic Crystal Structure Analysis of the 1,2-dihydroxynaphthalene dioxygeanse of Pseudomonas sp. strain C18 complexes to 1,2-dihydroxynaphthalene | | Descriptor: | 1,2-dihydroxynaphthalene dioxygenase, FE (II) ION, GLYCEROL, ... | | Authors: | Neau, D.B, Kelker, M.S, Colbert, C.L, Bolin, J.T. | | Deposit date: | 2007-03-10 | | Release date: | 2008-02-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural explanation for success and failure in the enzymatic ring-cleavage of 3,4 dihydroxybiphenyl and related PCB metabolites.
To be Published
|
|
2EHZ
 
 | | Anaerobic Crystal Structure Analysis of 1,2-dihydroxynaphthalene dioxygenase from Pseudomonas sp. strain C18 complexed with 4-methylcatechol | | Descriptor: | 1,2-ETHANEDIOL, 1,2-dihydroxynaphthalene dioxygenase, 4-METHYLCATECHOL, ... | | Authors: | Neau, D.B, Kelker, M.S, Colbert, C.L, Bolin, J.T. | | Deposit date: | 2007-03-10 | | Release date: | 2008-02-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural explanation for success and failure in the enzymatic ring-cleavage of 3,4 dihydroxybiphenyl and related PCB metabolites
To be Published
|
|
4QWT
 
 | |
6N2W
 
 | | The structure of Stable-5-Lipoxygenase bound to NDGA | | Descriptor: | 4-[(2R,3S)-3-[(3,4-DIHYDROXYPHENYL)METHYL]-2-METHYLBUTYL]BENZENE-1,2-DIOL, Arachidonate 5-lipoxygenase, FE (II) ION | | Authors: | Newcomer, M.E, Gilbert, N.C, Neau, D.B. | | Deposit date: | 2018-11-14 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural and mechanistic insights into 5-lipoxygenase inhibition by natural products.
Nat.Chem.Biol., 16, 2020
|
|
7LAF
 
 | | 15-lipoxygenase-2 loop mutant bound to imidazole-based inhibitor | | Descriptor: | 3-{[(4-methylphenyl)methyl]sulfanyl}-1-phenyl-1H-1,2,4-triazole, MANGANESE (II) ION, Polyunsaturated fatty acid lipoxygenase ALOX15B | | Authors: | Newcomer, M.E, Gilbert, N.C, Neau, D.B. | | Deposit date: | 2021-01-06 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Kinetic and structural investigations of novel inhibitors of human epithelial 15-lipoxygenase-2.
Bioorg.Med.Chem., 46, 2021
|
|
3O8Y
 
 | | Stable-5-Lipoxygenase | | Descriptor: | Arachidonate 5-lipoxygenase, FE (II) ION | | Authors: | Newcomer, M.E, Gilbert, N.C, Bartlett, S.G, Waight, M.T, Neau, D.B, Boeglin, W.E, Brash, A.R. | | Deposit date: | 2010-08-03 | | Release date: | 2011-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.389 Å) | | Cite: | The structure of human 5-lipoxygenase.
Science, 331, 2011
|
|
2ORX
 
 | | Structural Basis for Ligand Binding and Heparin Mediated Activation of Neuropilin | | Descriptor: | Neuropilin-1 | | Authors: | Vander Kooi, C.W, Jusino, M.A, Perman, B, Neau, D.B, Bellamy, H.D, Leahy, D.J. | | Deposit date: | 2007-02-05 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for ligand and heparin binding to neuropilin B domains
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2ORZ
 
 | | Structural Basis for Ligand Binding and Heparin Mediated Activation of Neuropilin | | Descriptor: | Neuropilin-1, Tuftsin | | Authors: | Vander Kooi, C.W, Jusino, M.A, Perman, B, Neau, D.B, Bellamy, H.D, Leahy, D.J. | | Deposit date: | 2007-02-05 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for ligand and heparin binding to neuropilin B domains.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4HR7
 
 | | Crystal Structure of Biotin Carboxyl Carrier Protein-Biotin Carboxylase Complex from E.coli | | Descriptor: | 1,2-ETHANEDIOL, Biotin carboxyl carrier protein of acetyl-CoA carboxylase, Biotin carboxylase, ... | | Authors: | Broussard, T.C, Kobe, M.J, Pakhomova, S, Neau, D.B, Price, A.E, Champion, T.S, Waldrop, G.L. | | Deposit date: | 2012-10-26 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | The three-dimensional structure of the biotin carboxylase-biotin carboxyl carrier protein complex of E. coli acetyl-CoA carboxylase.
Structure, 21, 2013
|
|
4RZQ
 
 | | Structural Analysis of Substrate, Reaction Intermediate and Product Binding in Haemophilus influenzae Biotin Carboxylase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Biotin carboxylase, methyl (3aS,4S,6aR)-4-(5-methoxy-5-oxopentyl)-2-oxohexahydro-1H-thieno[3,4-d]imidazole-1-carboxylate | | Authors: | Broussard, T.C, Pakhomova, S, Neau, D.B, Champion, T.S, Bonnot, R, Waldrop, G.L. | | Deposit date: | 2014-12-23 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Analysis of Substrate, Reaction Intermediate, and Product Binding in Haemophilus influenzae Biotin Carboxylase.
Biochemistry, 54, 2015
|
|
5DKR
 
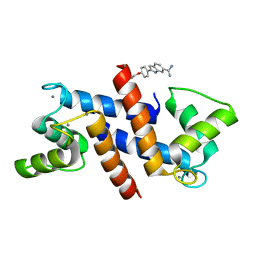 | | Crystal Structure of Calcium-loaded S100B bound to SBi29 | | Descriptor: | 2-[4-(4-carbamimidoylphenoxy)phenyl]-1H-indole-6-carboximidamide, CALCIUM ION, Protein S100-B | | Authors: | Cavalier, M.C, Ansari, M.I, Pierce, A.D, Wilder, P.T, McKnight, L.E, Raman, E.P, Neau, D.B, Bezawada, P, Alasady, M.J, Varney, K.M, Toth, E.A, MacKerell Jr, A.D, Coop, A, Weber, D.J. | | Deposit date: | 2015-09-03 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.742 Å) | | Cite: | Small Molecule Inhibitors of Ca(2+)-S100B Reveal Two Protein Conformations.
J.Med.Chem., 59, 2016
|
|
5DKQ
 
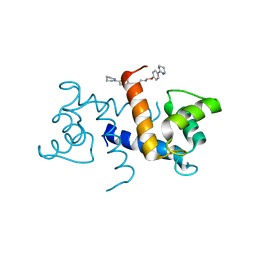 | | Crystal Structure of Calcium-loaded S100B bound to SBi4214 | | Descriptor: | 2,2'-[pentane-1,5-diylbis(oxybenzene-4,1-diyl)]di-1,4,5,6-tetrahydropyrimidine, CALCIUM ION, Protein S100-B | | Authors: | Cavalier, M.C, Ansari, M.I, Pierce, A.D, Wilder, P.T, McKnight, L.E, Raman, E.P, Neau, D.B, Bezawada, P, Alasady, M.J, Varney, K.M, Toth, E.A, MacKerell Jr, A.D, Coop, A, Weber, D.J. | | Deposit date: | 2015-09-03 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Small Molecule Inhibitors of Ca(2+)-S100B Reveal Two Protein Conformations.
J.Med.Chem., 59, 2016
|
|
5DKN
 
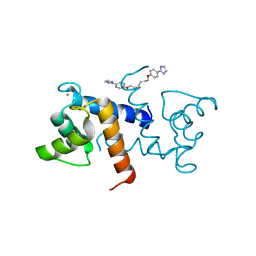 | | Crystal Structure of Calcium-loaded S100B bound to SBi4225 | | Descriptor: | 2,2'-[heptane-1,7-diylbis(oxybenzene-4,1-diyl)]bis(1H-imidazole), CALCIUM ION, Protein S100-B | | Authors: | Cavalier, M.C, Ansari, M.I, Pierce, A.D, Wilder, P.T, McKnight, L.E, Raman, E.P, Neau, D.B, Bezawada, P, Alasady, M.J, Varney, K.M, Toth, E.A, MacKerell Jr, A.D, Coop, A, Weber, D.J. | | Deposit date: | 2015-09-03 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.528 Å) | | Cite: | Small Molecule Inhibitors of Ca(2+)-S100B Reveal Two Protein Conformations.
J.Med.Chem., 59, 2016
|
|
6NCF
 
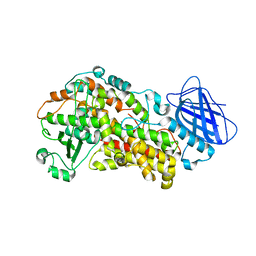 | | The structure of Stable-5-Lipoxygenase bound to AKBA | | Descriptor: | (3alpha,8alpha,17alpha,18alpha)-3-(acetyloxy)-11-oxours-12-en-23-oic acid, Arachidonate 5-lipoxygenase, FE (II) ION | | Authors: | Newcomer, M.E, Gilbert, N.C, Neau, D.B. | | Deposit date: | 2018-12-11 | | Release date: | 2020-05-13 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (2.871 Å) | | Cite: | Structural and mechanistic insights into 5-lipoxygenase inhibition by natural products.
Nat.Chem.Biol., 16, 2020
|
|
6NS6
 
 | | Crystal structure of fungal lipoxygenase from Fusarium graminearum. P21 crystal form. | | Descriptor: | FE (II) ION, lipoxygenase | | Authors: | Pakhomova, S, Boeglin, W.E, Neau, D.B, Bartlett, S.G, Brash, A.R, Newcomer, M.E. | | Deposit date: | 2019-01-24 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | An ensemble of lipoxygenase structures reveals novel conformations of the Fe coordination sphere.
Protein Sci., 28, 2019
|
|
6NS5
 
 | | Crystal structure of fungal lipoxygenase from Fusarium graminearum. Second C2 crystal form. | | Descriptor: | FE (II) ION, lipoxygenase | | Authors: | Pakhomova, S, Boeglin, W.E, Neau, D.B, Bartlett, S.G, Brash, A.R, Newcomer, M.E. | | Deposit date: | 2019-01-24 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | An ensemble of lipoxygenase structures reveals novel conformations of the Fe coordination sphere.
Protein Sci., 28, 2019
|
|
6NS2
 
 | | Crystal structure of fungal lipoxygenase from Fusarium graminearum. P212121 crystal form. | | Descriptor: | FE (II) ION, lipoxygenase | | Authors: | Pakhomova, S, Boeglin, W.E, Neau, D.B, Bartlett, S.G, Brash, A.R, Newcomer, M.E. | | Deposit date: | 2019-01-24 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | An ensemble of lipoxygenase structures reveals novel conformations of the Fe coordination sphere.
Protein Sci., 28, 2019
|
|
6NS3
 
 | | Crystal structure of fungal lipoxygenase from Fusarium graminearum. I222 crystal form. | | Descriptor: | FE (II) ION, lipoxygenase | | Authors: | Pakhomova, S, Boeglin, W.E, Neau, D.B, Bartlett, S.G, Brash, A.R, Newcomer, M.E. | | Deposit date: | 2019-01-24 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | An ensemble of lipoxygenase structures reveals novel conformations of the Fe coordination sphere.
Protein Sci., 28, 2019
|
|
6NS4
 
 | | Crystal structure of fungal lipoxygenase from Fusarium graminearum. C2 crystal form. | | Descriptor: | ACETATE ION, FE (II) ION, GLYCEROL, ... | | Authors: | Pakhomova, S, Boeglin, W.E, Neau, D.B, Bartlett, S.G, Brash, A.R, Newcomer, M.E. | | Deposit date: | 2019-01-24 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An ensemble of lipoxygenase structures reveals novel conformations of the Fe coordination sphere.
Protein Sci., 28, 2019
|
|
