4ANI
 
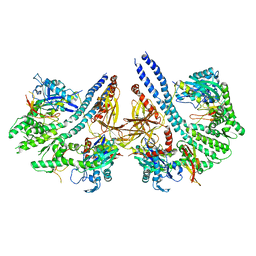 | | Structural basis for the intermolecular communication between DnaK and GrpE in the DnaK chaperone system from Geobacillus kaustophilus HTA426 | | Descriptor: | CHAPERONE PROTEIN DNAK, PROTEIN GRPE | | Authors: | Wu, C.-C, Naveen, V, Chien, C.-H, Chang, Y.-W, Hsiao, C.-D. | | Deposit date: | 2012-03-19 | | Release date: | 2012-05-23 | | Last modified: | 2012-08-01 | | Method: | X-RAY DIFFRACTION (4.094 Å) | | Cite: | Crystal Structure of Dnak Protein Complexed with Nucleotide Exchange Factor Grpe in Dnak Chaperone System: Insight Into Intermolecular Communication.
J.Biol.Chem., 287, 2012
|
|
5EZ1
 
 | |
5WTN
 
 | |
5BRQ
 
 | |
5BRP
 
 | |
