3ALP
 
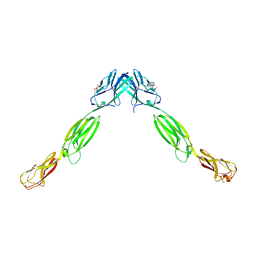 | | Cell adhesion protein | | Descriptor: | CITRIC ACID, HEXANE-1,6-DIOL, Poliovirus receptor-related protein 1 | | Authors: | Narita, H, Nakagawa, A, Suzuki, M. | | Deposit date: | 2010-08-05 | | Release date: | 2011-02-16 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Crystal Structure of the cis-Dimer of Nectin-1: implications for the architecture of cell-cell junctions
J.Biol.Chem., 286, 2011
|
|
7WJT
 
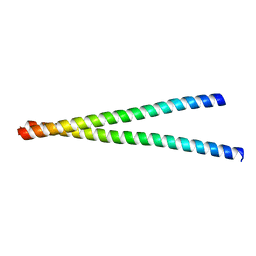 | |
1DHY
 
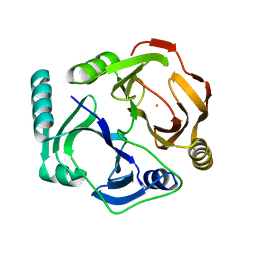 | | KKS102 BPHC ENZYME | | Descriptor: | 2,3-DIHYDROXYBIPHENYL 1,2-DIOXYGENASE, FE (III) ION | | Authors: | Senda, T, Sugiyama, K, Narita, H, Mitsui, Y. | | Deposit date: | 1995-07-07 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structures of free form and two substrate complexes of an extradiol ring-cleavage type dioxygenase, the BphC enzyme from Pseudomonas sp. strain KKS102.
J.Mol.Biol., 255, 1996
|
|
5B22
 
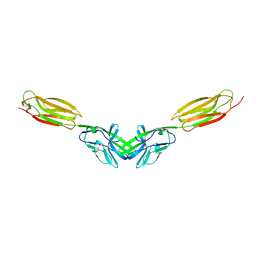 | | Dimer structure of murine Nectin-3 D1D2 | | Descriptor: | Nectin-3, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Takebe, K, Sangawa, T, Katsutani, T, Narita, H, Suzuki, M. | | Deposit date: | 2015-12-28 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Dimer structure of murine Nectin-3 D1D2
To Be Published
|
|
3VQF
 
 | | Crystal Structure Analysis of the PDZ Domain Derived from the Tight Junction Regulating Protein | | Descriptor: | E3 ubiquitin-protein ligase LNX | | Authors: | Akiyoshi, Y, Hamada, D, Goda, N, Tenno, T, Narita, H, Nakagawa, A, Furuse, M, Suzuki, M, Hiroaki, H. | | Deposit date: | 2012-03-22 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | Structural basis for down regulation of tight junction by PDZ-domain containing E3-Ubiquitin ligase
To be Published
|
|
3VQG
 
 | | Crystal Structure Analysis of the PDZ Domain Derived from the Tight Junction Regulating Protein | | Descriptor: | C-terminal peptide from Immunoglobulin superfamily member 5, E3 ubiquitin-protein ligase LNX, SULFATE ION | | Authors: | Akiyoshi, Y, Hamada, D, Goda, N, Tenno, T, Narita, H, Nakagawa, A, Furuse, M, Suzuki, M, Hiroaki, H. | | Deposit date: | 2012-03-23 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural basis for down regulation of tight junction by PDZ-domain containing E3-Ubiquitin ligase
To be Published
|
|
1V6Q
 
 | | Crystal Structures of Collagen Model Peptides with Pro-Hyp-Gly Sequence at 1.3 A | | Descriptor: | Collagen like peptide | | Authors: | Okuyama, K, Hongo, C, Fukushima, R, Wu, G, Narita, H, Noguchi, K, Tanaka, Y, Nishino, N. | | Deposit date: | 2003-12-03 | | Release date: | 2004-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structures of collagen model peptides with Pro-Hyp-Gly repeating sequence at 1.26 A resolution: implications for proline ring puckering
Biopolymers, 76, 2004
|
|
1V7H
 
 | | Crystal Structures of Collagen Model Peptides with Pro-Hyp-Gly Sequence at 1.26 A | | Descriptor: | Collagen like peptide | | Authors: | Okuyama, K, Hongo, C, Fukushima, R, Wu, G, Narita, H, Noguchi, K, Tanaka, Y, Nishino, N. | | Deposit date: | 2003-12-17 | | Release date: | 2004-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structures of collagen model peptides with Pro-Hyp-Gly repeating sequence at 1.26 A resolution: implications for proline ring puckering
Biopolymers, 76, 2004
|
|
3WKV
 
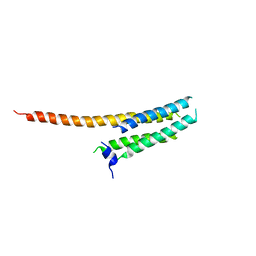 | | Voltage-gated proton channel: VSOP/Hv1 chimeric channel | | Descriptor: | Ion channel | | Authors: | Takeshita, K, Sakata, S, Yamashita, E, Fujiwara, Y, Kawanabe, A, Kurokawa, T, Okochi, Y, Matsuda, M, Narita, H, Okamura, Y, Nakagawa, A. | | Deposit date: | 2013-10-31 | | Release date: | 2014-03-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.453 Å) | | Cite: | X-ray crystal structure of voltage-gated proton channel.
Nat.Struct.Mol.Biol., 21, 2014
|
|
7DAA
 
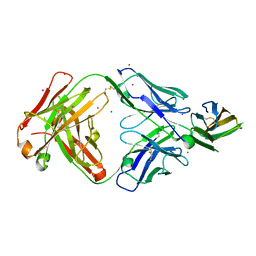 | | Crystal structure of basigin complexed with anti-basigin Fab fragment | | Descriptor: | CADMIUM ION, Heavy chain of antibody Fab fragment, Isoform 2 of Basigin, ... | | Authors: | Sakuragi, T, Kanai, R, Narita, H, Onishi, E, Miyazaki, T, Baba, T, Nakagawa, A, Toyoshima, C, Nagata, S. | | Deposit date: | 2020-10-16 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | The tertiary structure of the human Xkr8-Basigin complex that scrambles phospholipids at plasma membranes.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7DCE
 
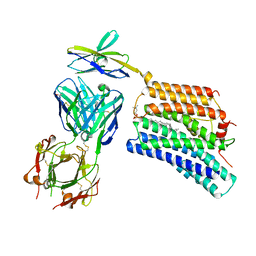 | | Cryo-EM structure of human XKR8-basigin complex bound to Fab fragment | | Descriptor: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Heavy chain of Fab fragment, Isoform 2 of Basigin, ... | | Authors: | Sakuragi, T, Kanai, R, Tsutsumi, A, Narita, H, Onishi, E, Miyazaki, T, Baba, T, Nakagawa, A, Kikkawa, M, Toyoshima, C, Nagata, S. | | Deposit date: | 2020-10-26 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The tertiary structure of the human Xkr8-Basigin complex that scrambles phospholipids at plasma membranes.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7D9Z
 
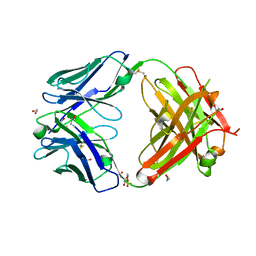 | | Crystal structure of anti-basigin Fab fragment | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, Heavy chain of antibody Fab fragment, ... | | Authors: | Sakuragi, T, Kanai, R, Narita, H, Onishi, E, Miyazaki, T, Baba, T, Nakagawa, A, Toyoshima, C, Nagata, S. | | Deposit date: | 2020-10-14 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.123 Å) | | Cite: | The tertiary structure of the human Xkr8-Basigin complex that scrambles phospholipids at plasma membranes.
Nat.Struct.Mol.Biol., 28, 2021
|
|
3AXA
 
 | | Crystal structure of afadin PDZ domain in complex with the C-terminal peptide from nectin-3 | | Descriptor: | Afadin, Nectin-3 | | Authors: | Fujiwara, Y, Goda, N, Narita, H, Satomura, K, Nakagawa, A, Sakisaka, T, Suzuki, M, Hiroaki, H. | | Deposit date: | 2011-03-31 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structure of afadin PDZ domain-nectin-3 complex shows the structural plasticity of the ligand-binding site.
Protein Sci., 24, 2015
|
|
3AV4
 
 | | Crystal structure of mouse DNA methyltransferase 1 | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, ZINC ION | | Authors: | Takeshita, K, Suetake, I, Yamashita, E, Suga, M, Narita, H, Nakagawa, A, Tajima, S. | | Deposit date: | 2011-02-22 | | Release date: | 2011-05-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural insight into maintenance methylation by mouse DNA methyltransferase 1 (Dnmt1).
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3AV5
 
 | | Crystal structure of mouse DNA methyltransferase 1 with AdoHcy | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Takeshita, K, Suetake, I, Yamashita, E, Suga, M, Narita, H, Nakagawa, A, Tajima, S. | | Deposit date: | 2011-02-22 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural insight into maintenance methylation by mouse DNA methyltransferase 1 (Dnmt1).
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3AV6
 
 | | Crystal structure of mouse DNA methyltransferase 1 with AdoMet | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, S-ADENOSYLMETHIONINE, ZINC ION | | Authors: | Takeshita, K, Suetake, I, Yamashita, E, Suga, M, Narita, H, Nakagawa, A, Tajima, S. | | Deposit date: | 2011-02-22 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structural insight into maintenance methylation by mouse DNA methyltransferase 1 (Dnmt1).
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2DRX
 
 | | Structure Analysis of (POG)4-(LOG)2-(POG)4 | | Descriptor: | collagen like peptide | | Authors: | Okuyama, K. | | Deposit date: | 2006-06-16 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Unique side chain conformation of a leu residue in a triple-helical structure
Biopolymers, 86, 2007
|
|
2DRT
 
 | | Structure Analysis of (POG)4-LOG-(POG)5 | | Descriptor: | ETHANOL, collagen like peptide | | Authors: | Okuyama, K. | | Deposit date: | 2006-06-14 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Unique side chain conformation of a leu residue in a triple-helical structure
Biopolymers, 86, 2007
|
|
1V4F
 
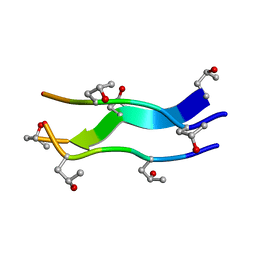 | | Crystal structures of collagen model peptides with pro-hyp-gly sequence at 1.3A | | Descriptor: | collagen like peptide | | Authors: | Okuyama, K, Hongo, C, Fukushima, R, Wu, G, Noguchi, K, Tanaka, Y, Nishino, N. | | Deposit date: | 2003-11-13 | | Release date: | 2004-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal structures of collagen model peptides with Pro-Hyp-Gly repeating sequence at 1.26 A resolution: implications for proline ring puckering
Biopolymers, 76, 2004
|
|
5X70
 
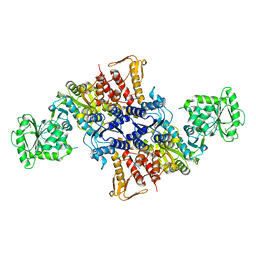 | |
5X6X
 
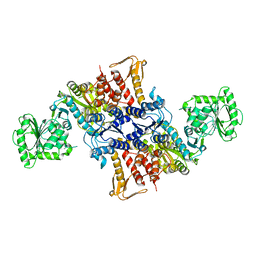 | |
5X6Z
 
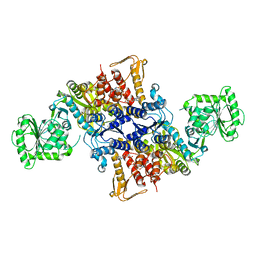 | |
5X6Y
 
 | |
5X71
 
 | |
1EIL
 
 | | 2,3-DIHYDROXYBIPHENYL-1,2-DIOXYGENASE | | Descriptor: | 2,3-DIHYDROXYBIPHENYL 1,2-DIOXYGENASE, FE (III) ION, SULFATE ION | | Authors: | Senda, T. | | Deposit date: | 2000-02-26 | | Release date: | 2001-02-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of substrate free and complex forms of reactivated BphC, an extradiol type ring-cleavage dioxygenase.
J.Inorg.Biochem., 83, 2001
|
|
