1J3A
 
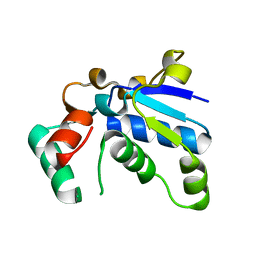 | | Crystal structure of ribosomal protein L13 from Pyrococcus horikoshii | | Descriptor: | 50S ribosomal protein L13P | | Authors: | Nakashima, T, Tanaka, M, Kazama, T, Kawamura, S, Kimura, M, Yao, M, Tanaka, I. | | Deposit date: | 2003-01-21 | | Release date: | 2003-02-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of ribosomal protein L13 from hyperthermophilic archaeon Pyrococcus horikoshii
To be Published
|
|
1IQ4
 
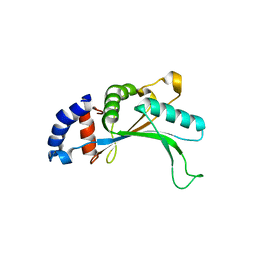 | | 5S-RRNA BINDING RIBOSOMAL PROTEIN L5 FROM BACILLUS STEAROTHERMOPHILUS | | Descriptor: | 50S RIBOSOMAL PROTEIN L5 | | Authors: | Nakashima, T, Yao, M, Kawamura, S, Iwasaki, K, Kimura, M, Tanaka, I. | | Deposit date: | 2001-06-13 | | Release date: | 2001-06-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ribosomal protein L5 has a highly twisted concave surface and flexible arms responsible for rRNA binding.
RNA, 7, 2001
|
|
2Z41
 
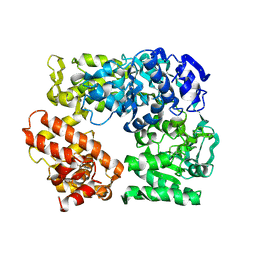 | | Crystal Structure Analysis of the Ski2-type RNA helicase | | Descriptor: | MAGNESIUM ION, putative ski2-type helicase | | Authors: | Nakashima, T, Zhang, X, Kakuta, Y, Yao, M, Tanaka, I, Kimura, M. | | Deposit date: | 2007-06-12 | | Release date: | 2008-04-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Crystal structure of an archaeal Ski2p-like protein from Pyrococcus horikoshii OT3
Protein Sci., 17, 2008
|
|
1RL2
 
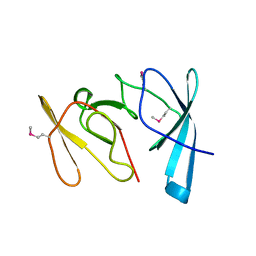 | |
2CZV
 
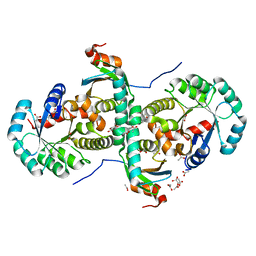 | | Crystal structure of archeal RNase P protein ph1481p in complex with ph1877p | | Descriptor: | ACETIC ACID, Ribonuclease P protein component 2, Ribonuclease P protein component 3, ... | | Authors: | Kawano, S, Kakuta, Y, Nakashima, T, Tanaka, I, Kimura, M. | | Deposit date: | 2005-07-19 | | Release date: | 2006-06-27 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of protein Ph1481p in complex with protein Ph1877p of archaeal RNase P from Pyrococcus horikoshii OT3: implication of dimer formation of the holoenzyme
J.Mol.Biol., 357, 2006
|
|
2Z7C
 
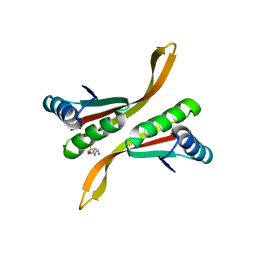 | | Crystal structure of chromatin protein alba from hyperthermophilic archaeon pyrococcus horikoshii | | Descriptor: | ARGININE, DNA/RNA-binding protein Alba | | Authors: | Hada, K, Nakashima, T, Osawa, T, Shimada, H, Kakuta, Y, Kimura, M. | | Deposit date: | 2007-08-17 | | Release date: | 2008-08-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure and functional analysis of an archaeal chromatin protein Alba from the hyperthermophilic archaeon Pyrococcus horikoshii OT3.
Biosci.Biotechnol.Biochem., 72, 2008
|
|
3WZ0
 
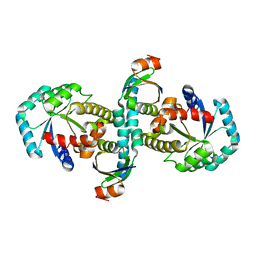 | | On archaeal homologs of the human RNase P proteins Pop5 and Rpp30 in the hyperthermophilic archaeon Thermococcus kodakarensis | | Descriptor: | Ribonuclease P protein component 2, Ribonuclease P protein component 3 | | Authors: | Suematsu, K, Ueda, T, Nakashima, T, Kakuta, Y, Kimura, M. | | Deposit date: | 2014-09-11 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | On archaeal homologs of the human RNase P proteins Pop5 and Rpp30 in the hyperthermophilic archaeon Thermococcus kodakarensis.
Biosci.Biotechnol.Biochem., 79, 2015
|
|
3WYZ
 
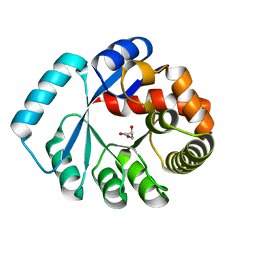 | | On archaeal homologs of the human RNase P protein Rpp30 in the hyperthermophilic archaeon Thermococcus kodakarensis | | Descriptor: | GLYCEROL, Ribonuclease P protein component 3 | | Authors: | Suematsu, K, Ueda, T, Nakashima, T, Kakuta, Y, Kimura, M. | | Deposit date: | 2014-09-11 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | On archaeal homologs of the human RNase P proteins Pop5 and Rpp30 in the hyperthermophilic archaeon Thermococcus kodakarensis.
Biosci.Biotechnol.Biochem., 79, 2015
|
|
1WNU
 
 | | Structure of Archaeal Trans-Editing Protein AlaX in complex with L-serine | | Descriptor: | SERINE, ZINC ION, alanyl-tRNA synthetase | | Authors: | Sokabe, M, Okada, A, Nakashima, T, Yao, M, Tanaka, I. | | Deposit date: | 2004-08-09 | | Release date: | 2005-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis of alanine discrimination in editing site
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1WXO
 
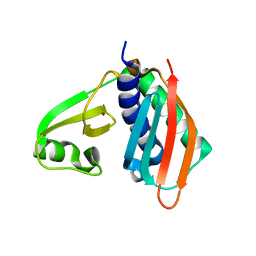 | | Structure of Archaeal Trans-Editing Protein AlaX in complex with zinc | | Descriptor: | ZINC ION, alanyl-tRNA synthetase | | Authors: | Sokabe, M, Okada, A, Nakashima, T, Yao, M, Tanaka, I. | | Deposit date: | 2005-01-27 | | Release date: | 2005-07-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Molecular basis of alanine discrimination in editing site
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
3AY8
 
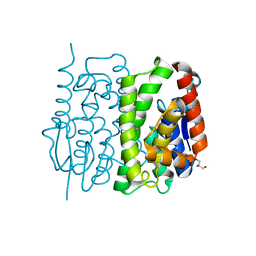 | | Glutathione S-transferase unclassified 2 from Bombyx mori | | Descriptor: | GLYCEROL, Glutathione S-transferase | | Authors: | Kakuta, Y, Usuda, K, Nakashima, T, Kimura, M, Aso, Y, Yamamoto, K. | | Deposit date: | 2011-05-02 | | Release date: | 2011-09-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic survey of active sites of an unclassified glutathione transferase from Bombyx mori
Biochim.Biophys.Acta, 1810, 2011
|
|
1BK7
 
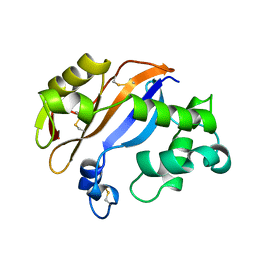 | | RIBONUCLEASE MC1 FROM THE SEEDS OF BITTER GOURD | | Descriptor: | PROTEIN (RIBONUCLEASE MC1) | | Authors: | Nakagawa, A, Tanaka, I. | | Deposit date: | 1998-07-15 | | Release date: | 1999-07-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a ribonuclease from the seeds of bitter gourd (Momordica charantia) at 1.75 A resolution.
Biochim.Biophys.Acta, 1433, 1999
|
|
3GP7
 
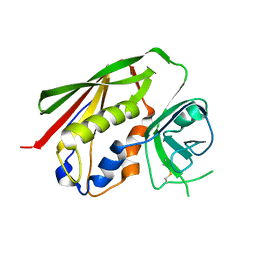 | |
5DCV
 
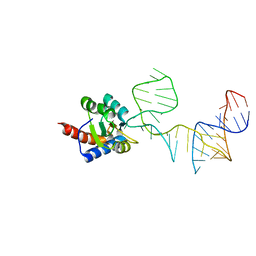 | | Crystal structure of PhoRpp38-SL12M complex | | Descriptor: | 50S ribosomal protein L7Ae, RNA (47-MER) | | Authors: | Oshima, K, Tanaka, Y, Yao, M. | | Deposit date: | 2015-08-24 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.401 Å) | | Cite: | Structural basis for recognition of a kink-turn motif by an archaeal homologue of human RNase P protein Rpp38
Biochem.Biophys.Res.Commun., 474, 2016
|
|
5XTM
 
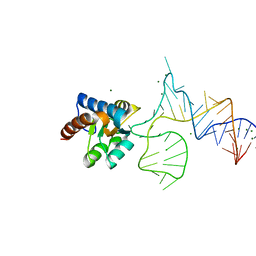 | | Crystal structure of PhoRpp38 bound to a K-turn in P12.2 helix | | Descriptor: | 50S ribosomal protein L7Ae, MAGNESIUM ION, RNA (47-MER) | | Authors: | Oshima, K, Kimura, M. | | Deposit date: | 2017-06-20 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the archaeal RNase P protein Rpp38 in complex with RNA fragments containing a K-turn motif.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5Y7M
 
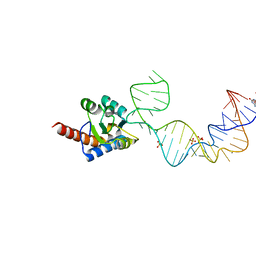 | | Crystal structure of PhoRpp38 bound to a K-turn in P12.1 helix | | Descriptor: | 50S ribosomal protein L7Ae, GUANOSINE-5'-TRIPHOSPHATE, RNA (52-MER), ... | | Authors: | Oshima, K, Kimura, M. | | Deposit date: | 2017-08-01 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of the archaeal RNase P protein Rpp38 in complex with RNA fragments containing a K-turn motif.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
3AML
 
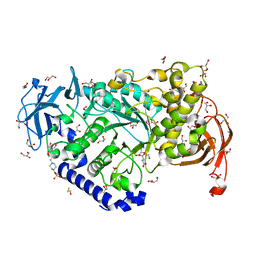 | | Structure of the Starch Branching Enzyme I (BEI) from Oryza sativa L | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Kakuta, Y, Chaen, K, Noguchi, J, Vu, N, Kimura, M. | | Deposit date: | 2010-08-20 | | Release date: | 2011-09-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the branching enzyme I (BEI) from Oryza sativa L with implications for catalysis and substrate binding.
Glycobiology, 21, 2011
|
|
3AMK
 
 | | Structure of the Starch Branching Enzyme I (BEI) from Oryza sativa L | | Descriptor: | GLYCEROL, Os06g0726400 protein, PHOSPHATE ION | | Authors: | Kakuta, Y, Chaen, K, Noguchi, J, Vu, N, Kimura, M. | | Deposit date: | 2010-08-20 | | Release date: | 2011-09-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the branching enzyme I (BEI) from Oryza sativa L with implications for catalysis and substrate binding.
Glycobiology, 21, 2011
|
|
1J1G
 
 | | Crystal structure of the RNase MC1 mutant N71S in complex with 5'-GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Ribonuclease MC1 | | Authors: | Numata, T, Suzuki, A, Kakuta, Y, Kimura, K, Yao, M, Tanaka, I, Yoshida, Y, Ueda, T, Kimura, M. | | Deposit date: | 2002-12-04 | | Release date: | 2003-05-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of the Ribonuclease MC1 Mutants N71T and N71S in Complex with 5'-GMP: Structural Basis for Alterations in Substrate Specificity
Biochemistry, 42, 2003
|
|
1J1F
 
 | | Crystal structure of the RNase MC1 mutant N71T in complex with 5'-GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, RIBONUCLEASE MC1 | | Authors: | Numata, T, Suzuki, A, Kakuta, Y, Kimura, K, Yao, M, Tanaka, I, Yoshida, Y, Ueda, T, Kimura, M. | | Deposit date: | 2002-12-03 | | Release date: | 2003-05-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of the Ribonuclease MC1 Mutants N71T and N71S in Complex with 5'-GMP: Structural Basis for Alterations in Substrate Specificity
Biochemistry, 42, 2003
|
|
1V7O
 
 | |
487D
 
 | |
