4F1N
 
 | | Crystal structure of Kluyveromyces polysporus Argonaute with a guide RNA | | Descriptor: | KpAGO, RNA 5'-R(P*UP*AP*AP*AP*AP*AP*AP*AP*A)-3' | | Authors: | Nakanishi, K, Weinberg, D.E, Bartel, D.P, Patel, D.J. | | Deposit date: | 2012-05-07 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.187 Å) | | Cite: | Structure of yeast Argonaute with guide RNA.
Nature, 486, 2012
|
|
3HJ7
 
 | | Crystal structure of TILS C-terminal domain | | Descriptor: | CHLORIDE ION, tRNA(Ile)-lysidine synthase | | Authors: | Nakanishi, K, Bonnefond, L, Kimura, S, Suzuki, T, Ishitani, R, Nureki, O. | | Deposit date: | 2009-05-21 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for translational fidelity ensured by transfer RNA lysidine synthetase.
Nature, 461, 2009
|
|
4KXT
 
 | |
3A2K
 
 | | Crystal structure of TilS complexed with tRNA | | Descriptor: | bacterial tRNA, tRNA(Ile)-lysidine synthase | | Authors: | Nakanishi, K, Bonnefond, L, Ishitani, R, Nureki, O. | | Deposit date: | 2009-05-23 | | Release date: | 2009-10-20 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Structural basis for translational fidelity ensured by transfer RNA lysidine synthetase.
Nature, 461, 2009
|
|
2CSX
 
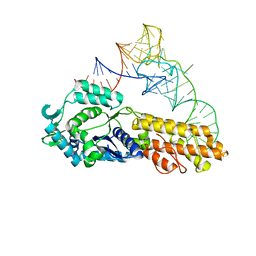 | | Crystal structure of Aquifex aeolicus methionyl-tRNA synthetase complexed with tRNA(Met) | | Descriptor: | Methionyl-tRNA synthetase, RNA (75-MER) | | Authors: | Nakanishi, K, Ogiso, Y, Nakama, T, Fukai, S, Nureki, O, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-23 | | Release date: | 2005-09-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for anticodon recognition by methionyl-tRNA synthetase.
Nat.Struct.Mol.Biol., 12, 2005
|
|
2CT8
 
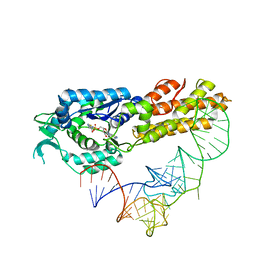 | | Crystal structure of Aquifex aeolicus methionyl-tRNA synthetase complexed with tRNA(Met) and methionyl-adenylate anologue | | Descriptor: | 5'-O-[(L-METHIONYL)-SULPHAMOYL]ADENOSINE, Methionyl-tRNA synthetase, RNA (74-MER) | | Authors: | Nakanishi, K, Ogiso, Y, Nakama, T, Fukai, S, Nureki, O, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-23 | | Release date: | 2005-09-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for anticodon recognition by methionyl-tRNA synthetase.
Nat.Struct.Mol.Biol., 12, 2005
|
|
1WY5
 
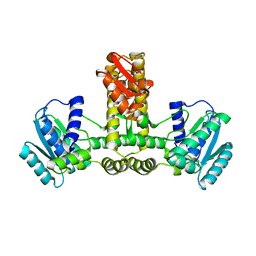 | | Crystal structure of isoluecyl-tRNA lysidine synthetase | | Descriptor: | Hypothetical UPF0072 protein AQ_1887 | | Authors: | Nakanishi, K, Fukai, S, Ikeuchi, Y, Soma, A, Sekine, Y, Suzuki, T, Nureki, O, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-02-06 | | Release date: | 2005-05-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural basis for lysidine formation by ATP pyrophosphatase accompanied by a lysine-specific loop and a tRNA-recognition domain.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
3RV1
 
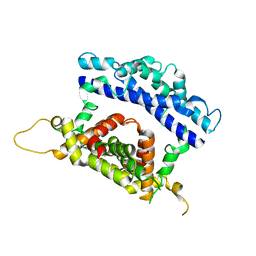 | | Crystal structure of the N-terminal and RNase III domains of K. polysporus Dcr1 E224Q mutant | | Descriptor: | K. polysporus Dcr1 | | Authors: | Nakanishi, K, Weinberg, D.E, Bartel, D.P, Patel, D.J. | | Deposit date: | 2011-05-05 | | Release date: | 2011-08-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.975 Å) | | Cite: | The inside-out mechanism of dicers from budding yeasts.
Cell(Cambridge,Mass.), 146, 2011
|
|
3RV0
 
 | | Crystal structure of K. polysporus Dcr1 without the C-terminal dsRBD | | Descriptor: | K. polysporus Dcr1, MAGNESIUM ION | | Authors: | Nakanishi, K, Weinberg, D.E, Bartel, D.P, Patel, D.J. | | Deposit date: | 2011-05-05 | | Release date: | 2011-08-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | The inside-out mechanism of dicers from budding yeasts.
Cell(Cambridge,Mass.), 146, 2011
|
|
6OON
 
 | | Human Argonaute4 bound to guide RNA | | Descriptor: | Protein argonaute-4, RNA (5'-R(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*UP*U)-3') | | Authors: | Park, M.S, Brackbill, J.A, Nakanishi, K. | | Deposit date: | 2019-04-23 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multidomain Convergence of Argonaute during RISC Assembly Correlates with the Formation of Internal Water Clusters.
Mol.Cell, 75, 2019
|
|
7KPW
 
 | | Structure of the H-lobe of yeast CKM | | Descriptor: | Mediator of RNA polymerase II transcription subunit 12 | | Authors: | Li, Y.C, Chao, T.C, Kim, H.J, Cholko, T, Chen, S.F, Nakanishi, K, Chang, C.E, Murakami, K, Garcia, B.A, Boyer, T.G, Tsai, K.L. | | Deposit date: | 2020-11-12 | | Release date: | 2021-01-27 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structure and noncanonical Cdk8 activation mechanism within an Argonaute-containing Mediator kinase module.
Sci Adv, 7, 2021
|
|
7KPX
 
 | | Structure of the yeast CKM | | Descriptor: | Mediator of RNA polymerase II transcription subunit 12, Mediator of RNA polymerase II transcription subunit 13, Meiotic mRNA stability protein kinase SSN3, ... | | Authors: | Li, Y.C, Chao, T.C, Kim, H.J, Cholko, T, Chen, S.F, Nakanishi, K, Chang, C.E, Murakami, K, Garcia, B.A, Boyer, T.G, Tsai, K.L. | | Deposit date: | 2020-11-12 | | Release date: | 2021-01-27 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure and noncanonical Cdk8 activation mechanism within an Argonaute-containing Mediator kinase module.
Sci Adv, 7, 2021
|
|
7KPV
 
 | | Structure of kinase and Central lobes of yeast CKM | | Descriptor: | Mediator of RNA polymerase II transcription subunit 12, Mediator of RNA polymerase II transcription subunit 13, Meiotic mRNA stability protein kinase SSN3, ... | | Authors: | Li, Y.C, Chao, T.C, Kim, H.J, Cholko, T, Chen, S.F, Nakanishi, K, Chang, C.E, Murakami, K, Garcia, B.A, Boyer, T.G, Tsai, K.L. | | Deposit date: | 2020-11-12 | | Release date: | 2021-01-27 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and noncanonical Cdk8 activation mechanism within an Argonaute-containing Mediator kinase module.
Sci Adv, 7, 2021
|
|
5VXB
 
 | |
5THE
 
 | |
5VM9
 
 | | Human Argonaute3 bound to guide RNA | | Descriptor: | Protein argonaute-3, RNA (5'-R(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*UP*U)-3'), RNA (5'-R(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*UP*U)-3') | | Authors: | Park, M.S, Nakanishi, K. | | Deposit date: | 2017-04-26 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Human Argonaute3 has slicer activity.
Nucleic Acids Res., 45, 2017
|
|
1TFO
 
 | | Ribonuclease from Escherichia coli complexed with its inhibitor protein | | Descriptor: | Colicin D, Colicin D immunity protein | | Authors: | Yajima, S, Nakanishi, K, Takahashi, K, Ogawa, T, Kezuka, Y, Hidaka, M, Nonaka, T, Ohsawa, K, Masaki, H. | | Deposit date: | 2004-05-27 | | Release date: | 2005-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Relation between tRNase activity and the structure of colicin D according to X-ray crystallography
Biochem.Biophys.Res.Commun., 322, 2004
|
|
1TFK
 
 | | Ribonuclease from Escherichia coli complexed with its inhibtor protein | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Colicin D, Colicin D immunity protein | | Authors: | Yajima, S, Nakanishi, K, Takahashi, K, Ogawa, T, Kezuka, Y, Hidaka, M, Nonaka, T, Ohsawa, K, Masaki, H. | | Deposit date: | 2004-05-27 | | Release date: | 2005-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Relation between tRNase activity and the structure of colicin D according to X-ray crystallography
Biochem.Biophys.Res.Commun., 322, 2004
|
|
3W4K
 
 | | Crystal Structure of human DAAO in complex with coumpound 13 | | Descriptor: | 3-hydroxy-6-(2-phenylethyl)pyridazin-4(1H)-one, D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hondo, T, Warizaya, M, Niimi, T, Namatame, I, Yamaguchi, T, Nakanishi, K, Hamajima, T, Harada, K, Sakashita, H, Matsumoto, Y, Orita, M, Watanabe, T, Takeuchi, M. | | Deposit date: | 2013-01-09 | | Release date: | 2013-05-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | 4-Hydroxypyridazin-3(2H)-one Derivatives as Novel d-Amino Acid Oxidase Inhibitors.
J.Med.Chem., 56, 2013
|
|
3W4J
 
 | | Crystal Structure of human DAAO in complex with coumpound 12 | | Descriptor: | 3-hydroxy-5-(2-phenylethyl)pyridin-2(1H)-one, D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hondo, T, Warizaya, M, Niimi, T, Namatame, I, Yamaguchi, T, Nakanishi, K, Hamajima, T, Harada, K, Sakashita, H, Matsumoto, Y, Orita, M, Watanabe, T, Takeuchi, M. | | Deposit date: | 2013-01-09 | | Release date: | 2013-05-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | 4-Hydroxypyridazin-3(2H)-one Derivatives as Novel d-Amino Acid Oxidase Inhibitors.
J.Med.Chem., 56, 2013
|
|
3W4I
 
 | | Crystal Structure of human DAAO in complex with coumpound 8 | | Descriptor: | D-amino-acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE, pyridine-2,3-diol | | Authors: | Hondo, T, Warizaya, M, Niimi, T, Namatame, I, Yamaguchi, T, Nakanishi, K, Hamajima, T, Harada, K, Sakashita, H, Matsumoto, Y, Orita, M, Watanabe, T, Takeuchi, M. | | Deposit date: | 2013-01-09 | | Release date: | 2013-05-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 4-Hydroxypyridazin-3(2H)-one Derivatives as Novel d-Amino Acid Oxidase Inhibitors.
J.Med.Chem., 56, 2013
|
|
8SQU
 
 | | Monomeric MapSPARTA bound with guide RNA and target DNA hybrid | | Descriptor: | MAGNESIUM ION, TIR-APAZ, guide RNA, ... | | Authors: | Shen, Z.F, Yang, X.Y, Fu, T.M. | | Deposit date: | 2023-05-04 | | Release date: | 2023-08-23 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Oligomerization-mediated activation of a short prokaryotic Argonaute.
Nature, 621, 2023
|
|
8SP3
 
 | | Asymmetric dimer of MapSPARTA bound with gRNA/tDNA hybrid | | Descriptor: | MAGNESIUM ION, TIR-APAZ, guide RNA, ... | | Authors: | Shen, Z.F, Yang, X.Y, Fu, T.M. | | Deposit date: | 2023-05-01 | | Release date: | 2023-08-23 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Oligomerization-mediated activation of a short prokaryotic Argonaute.
Nature, 621, 2023
|
|
8SP0
 
 | | Symmetric dimer of MapSPARTA bound with gRNA/tDNA hybrid | | Descriptor: | MAGNESIUM ION, TIR-APAZ, guide RNA, ... | | Authors: | Shen, Z.F, Yang, X.Y, Fu, T.M. | | Deposit date: | 2023-05-01 | | Release date: | 2023-08-23 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Oligomerization-mediated activation of a short prokaryotic Argonaute.
Nature, 621, 2023
|
|
8SPO
 
 | | Tetramerized activation of MapSPARTA bound with NAD+ | | Descriptor: | MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Piwi domain-containing protein, ... | | Authors: | Shen, Z.F, Yang, X.Y, Fu, T.M. | | Deposit date: | 2023-05-03 | | Release date: | 2023-08-23 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Oligomerization-mediated activation of a short prokaryotic Argonaute.
Nature, 621, 2023
|
|
