1FJM
 
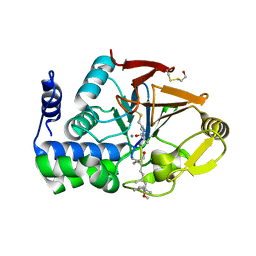 | | Protein serine/threonine phosphatase-1 (alpha isoform, type 1) complexed with microcystin-LR toxin | | Descriptor: | BETA-MERCAPTOETHANOL, MANGANESE (II) ION, PROTEIN SERINE/THREONINE PHOSPHATASE-1 (ALPHA ISOFORM, ... | | Authors: | Goldberg, J, Nairn, A.C, Kuriyan, J. | | Deposit date: | 1995-12-17 | | Release date: | 1996-06-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional structure of the catalytic subunit of protein serine/threonine phosphatase-1.
Nature, 376, 1995
|
|
1IAJ
 
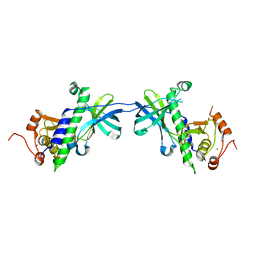 | | CRYSTAL STRUCTURE OF THE ATYPICAL PROTEIN KINASE DOMAIN OF A TRP CA-CHANNEL, CHAK (APO) | | Descriptor: | TRANSIENT RECEPTOR POTENTIAL-RELATED PROTEIN, ZINC ION | | Authors: | Yamaguchi, H, Matsushita, M, Nairn, A.C, Kuriyan, J. | | Deposit date: | 2001-03-22 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the atypical protein kinase domain of a TRP channel with phosphotransferase activity.
Mol.Cell, 7, 2001
|
|
1IAH
 
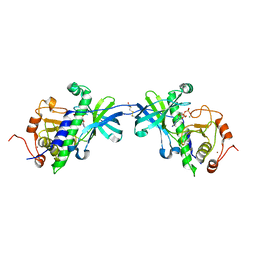 | | CRYSTAL STRUCTURE OF THE ATYPICAL PROTEIN KINASE DOMAIN OF A TRP CA-CHANNEL, CHAK (ADP-MG COMPLEX) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yamaguchi, H, Matsushita, M, Nairn, A.C, Kuriyan, J. | | Deposit date: | 2001-03-22 | | Release date: | 2001-06-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the atypical protein kinase domain of a TRP channel with phosphotransferase activity.
Mol.Cell, 7, 2001
|
|
1IA9
 
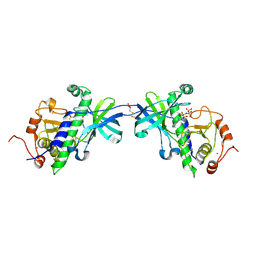 | | CRYSTAL STRUCTURE OF THE ATYPICAL PROTEIN KINASE DOMAIN OF A TRP CA-CHANNEL, CHAK (AMPPNP COMPLEX) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, TRANSIENT RECEPTOR POTENTIAL-RELATED PROTEIN, ... | | Authors: | Yamaguchi, H, Matsushita, M, Nairn, A.C, Kuriyan, J. | | Deposit date: | 2001-03-22 | | Release date: | 2001-06-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the atypical protein kinase domain of a TRP channel with phosphotransferase activity.
Mol.Cell, 7, 2001
|
|
1HKX
 
 | | Crystal structure of calcium/calmodulin-dependent protein kinase | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CALCIUM/CALMODULIN-DEPENDENT PROTEIN KINASE TYPE II ALPHA CHAIN, CHLORIDE ION, ... | | Authors: | Hoelz, A, Nairn, A.C, Kuriyan, J. | | Deposit date: | 2003-03-12 | | Release date: | 2003-06-02 | | Last modified: | 2023-01-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structure of a Tetradecameric Assembly of the Association Domain of Ca2+/Calmodulin-Dependent Kinase II
Mol.Cell, 11, 2003
|
|
1A06
 
 | | CALMODULIN-DEPENDENT PROTEIN KINASE FROM RAT | | Descriptor: | CALCIUM/CALMODULIN-DEPENDENT PROTEIN KINASE | | Authors: | Kuriyan, J, Goldberg, J. | | Deposit date: | 1997-12-09 | | Release date: | 1998-04-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the autoinhibition of calcium/calmodulin-dependent protein kinase I.
Cell(Cambridge,Mass.), 84, 1996
|
|
2BDW
 
 | |
3HVQ
 
 | | Crystal structure of a complex between Protein Phosphatase 1 alpha (PP1) and the PP1 binding and PDZ domains of Neurabin | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Neurabin-1, ... | | Authors: | Critton, D.A, Ragusa, M.J, Page, R, Peti, W. | | Deposit date: | 2009-06-16 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Spinophilin directs protein phosphatase 1 specificity by blocking substrate binding sites.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3EGG
 
 | | Crystal structure of a complex between Protein Phosphatase 1 alpha (PP1) and the PP1 binding and PDZ domains of Spinophilin | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Ragusa, M.J, Page, R, Peti, W. | | Deposit date: | 2008-09-10 | | Release date: | 2010-03-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Spinophilin directs protein phosphatase 1 specificity by blocking substrate binding sites.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3EGH
 
 | | Crystal structure of a complex between Protein Phosphatase 1 alpha (PP1), the PP1 binding and PDZ domains of Spinophilin and the small natural molecular toxin Nodularin-R | | Descriptor: | GLYCEROL, MANGANESE (II) ION, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit, ... | | Authors: | Ragusa, M.J, Page, R, Peti, W. | | Deposit date: | 2008-09-10 | | Release date: | 2010-03-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Spinophilin directs protein phosphatase 1 specificity by blocking substrate binding sites.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2F86
 
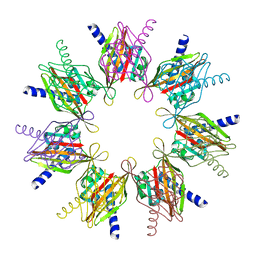 | |
2G5M
 
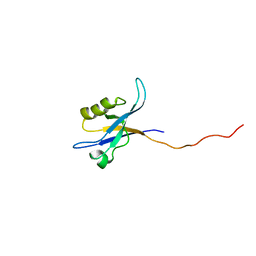 | | Spinophilin PDZ domain | | Descriptor: | Neurabin-2 | | Authors: | Kelker, M.S, Peti, W. | | Deposit date: | 2006-02-23 | | Release date: | 2007-01-09 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structural basis for spinophilin-neurabin receptor interaction.
Biochemistry, 46, 2007
|
|
2FN5
 
 | |
2GLE
 
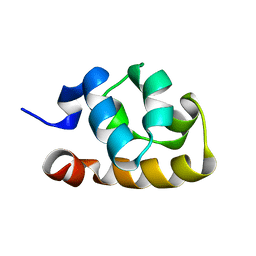 | |
1CDK
 
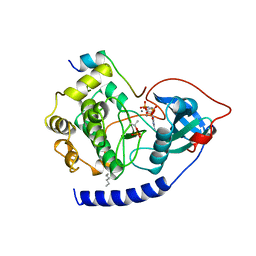 | | CAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT (E.C.2.7.1.37) (PROTEIN KINASE A) COMPLEXED WITH PROTEIN KINASE INHIBITOR PEPTIDE FRAGMENT 5-24 (PKI(5-24) ISOELECTRIC VARIANT CA) AND MN2+ ADENYLYL IMIDODIPHOSPHATE (MNAMP-PNP) AT PH 5.6 AND 7C AND 4C | | Descriptor: | CAMP-DEPENDENT PROTEIN KINASE, MANGANESE (II) ION, MYRISTIC ACID, ... | | Authors: | Bossemeyer, D, Engh, R.A, Kinzel, V, Ponstingl, H, Huber, R. | | Deposit date: | 1994-07-04 | | Release date: | 1995-10-15 | | Last modified: | 2012-07-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphotransferase and substrate binding mechanism of the cAMP-dependent protein kinase catalytic subunit from porcine heart as deduced from the 2.0 A structure of the complex with Mn2+ adenylyl imidodiphosphate and inhibitor peptide PKI(5-24).
EMBO J., 12, 1993
|
|
4MOV
 
 | | 1.45 A Resolution Crystal Structure of Protein Phosphatase 1 | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4503 Å) | | Cite: | Understanding the antagonism of retinoblastoma protein dephosphorylation by PNUTS provides insights into the PP1 regulatory code.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4MOY
 
 | | Structure of a second nuclear PP1 Holoenzyme, crystal form 1 | | Descriptor: | CHLORIDE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Choy, M.S, Hieke, M, Peti, W, Page, R. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1953 Å) | | Cite: | Understanding the antagonism of retinoblastoma protein dephosphorylation by PNUTS provides insights into the PP1 regulatory code.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4MP0
 
 | | Structure of a second nuclear PP1 Holoenzyme, crystal form 2 | | Descriptor: | GLYCEROL, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Choy, M.S, Hieke, M, Peti, W, Page, R. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1003 Å) | | Cite: | Understanding the antagonism of retinoblastoma protein dephosphorylation by PNUTS provides insights into the PP1 regulatory code.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5OVR
 
 | | X-Ray Characterization of Striatal-Enriched Protein Tyrosine Phosphatase Inhibitors | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 5, [(~{S})-[4-[3-[(~{R})-(3,4-dichlorophenyl)-oxidanyl-methyl]phenyl]phenyl]-oxidanyl-methyl]phosphonic acid | | Authors: | Kack, H, Wissler, L. | | Deposit date: | 2017-08-29 | | Release date: | 2017-11-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | X-ray Characterization and Structure-Based Optimization of Striatal-Enriched Protein Tyrosine Phosphatase Inhibitors.
J. Med. Chem., 60, 2017
|
|
5OVX
 
 | | X-Ray Characterization of Striatal-Enriched Protein Tyrosine Phosphatase Inhibitors | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 5, [(~{S})-[4-[3-[(~{S})-(3,4-dichlorophenyl)-oxidanyl-methyl]phenyl]phenyl]-oxidanyl-methyl]phosphonic acid | | Authors: | Kack, H, Wissler, L. | | Deposit date: | 2017-08-30 | | Release date: | 2017-11-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray Characterization and Structure-Based Optimization of Striatal-Enriched Protein Tyrosine Phosphatase Inhibitors.
J. Med. Chem., 60, 2017
|
|
5OW1
 
 | |
1EVB
 
 | |
1EVA
 
 | |
1LCM
 
 | |
