3CEZ
 
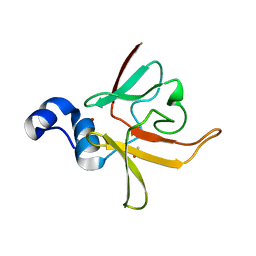 | | Crystal structure of methionine-R-sulfoxide reductase from Burkholderia pseudomallei | | Descriptor: | ACETIC ACID, Methionine-R-sulfoxide reductase, ZINC ION | | Authors: | Staker, B, Napuli, A, Nakazawa, S.H, Castaneda, L, Alkafeef, S, Vanvoorhis, W, Stewart, L, Myler, P, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2008-02-29 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Methionine-R-sulfoxide reductase from Burkholderia pseudomallei.
To be Published
|
|
4W5K
 
 | |
6O0A
 
 | |
1TQX
 
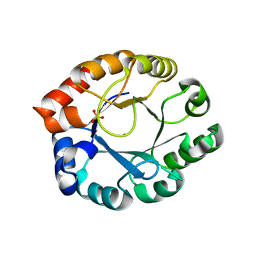 | | Crystal Structure of Pfal009167 A Putative D-Ribulose 5-Phosphate 3-Epimerase from P.falciparum | | Descriptor: | D-ribulose-5-phosphate 3-epimerase, putative, SULFATE ION, ... | | Authors: | Caruthers, J, Bosch, J, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2004-06-18 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a ribulose 5-phosphate 3-epimerase from Plasmodium falciparum.
Proteins, 62, 2006
|
|
3HGB
 
 | |
2B4R
 
 | | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase from Plasmodium falciparum at 2.25 Angstrom Resolution reveals intriguing extra electron density in the active site | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Robien, M.A, Bosch, J, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2005-09-26 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase from Plasmodium falciparum at 2.25 A resolution reveals intriguing extra electron density in the active site
Proteins, 62, 2006
|
|
2B4T
 
 | | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase from Plasmodium falciparum at 2.25 Angstrom resolution reveals intriguing extra electron density in the active site | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Robien, M.A, Bosch, J, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2005-09-26 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase from Plasmodium falciparum at 2.25 A resolution reveals intriguing extra electron density in the active site
Proteins, 62, 2006
|
|
5KOB
 
 | |
3IFT
 
 | | Crystal structure of glycine cleavage system protein H from Mycobacterium tuberculosis, using X-rays from the Compact Light Source. | | Descriptor: | Glycine cleavage system H protein | | Authors: | Edwards, T.E, Abendroth, J, Staker, B, Mayer, C, Phan, I, Kelley, A, Analau, E, Leibly, D, Rifkin, J, Loewen, R, Ruth, R.D, Stewart, L.J, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2009-07-25 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure determination of the glycine cleavage system protein H of Mycobacterium tuberculosis using an inverse Compton synchrotron X-ray source.
J.Struct.Funct.Genom., 11, 2010
|
|
4H51
 
 | |
4H7P
 
 | |
4I1I
 
 | |
3LR3
 
 | |
3LR0
 
 | |
3LR4
 
 | |
3LR5
 
 | |
4EFF
 
 | |
4EU1
 
 | |
4F4E
 
 | |
4F3P
 
 | |
3MEB
 
 | |
