2VZ5
 
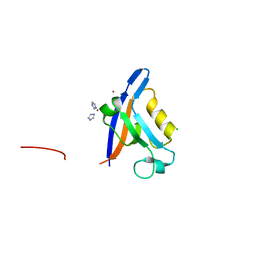 | | Structure of the PDZ domain of Tax1 (human T-cell leukemia virus type I) binding protein 3 | | Descriptor: | CHLORIDE ION, IMIDAZOLE, TAX1-BINDING PROTEIN 3, ... | | Authors: | Murray, J.W, Shafqat, N, Yue, W, Pilka, E, Johannsson, C, Salah, E, Cooper, C, Elkins, J.M, Pike, A.C, Roos, A, Filippakopoulos, P, von Delft, F, Wickstroem, M, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Oppermann, U. | | Deposit date: | 2008-07-30 | | Release date: | 2008-08-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.738 Å) | | Cite: | The Structure of the Pdz Domain of Tax1BP
To be Published
|
|
2VSW
 
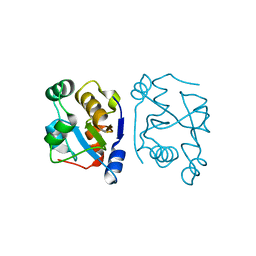 | | The structure of the rhodanese domain of the human dual specificity phosphatase 16 | | Descriptor: | DUAL SPECIFICITY PROTEIN PHOSPHATASE 16 | | Authors: | Murray, J.W, Barr, A, Pike, A.C.W, Elkins, J, Phillips, C, Wang, J, Savitsky, P, Roos, A, Bishop, S, Wickstroem, M, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Burgess-Brown, N, Pantic, N, Bray, J, von Delft, F, Gileadi, O, Knapp, S. | | Deposit date: | 2008-04-30 | | Release date: | 2008-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of the Rhodanese Domain of the Human Dual Specifity Phosphatase 16
To be Published
|
|
3I3N
 
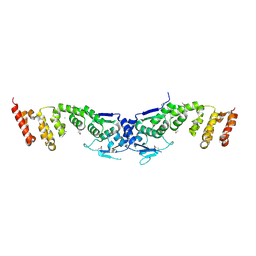 | | Crystal structure of the BTB-BACK domains of human KLHL11 | | Descriptor: | CHLORIDE ION, Kelch-like protein 11, THIOCYANATE ION | | Authors: | Murray, J.W, Cooper, C.D.O, Krojer, T, Mahajan, P, Salah, E, Keates, T, Savitsky, P, Pike, A.C.W, Roos, A, Muniz, J, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Knapp, S, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-06-30 | | Release date: | 2009-08-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the BTB-BACK domains of human KLHL11
To be Published
|
|
2BNL
 
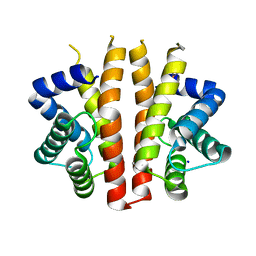 | |
2VJT
 
 | |
2VJH
 
 | | The structure of Phycoerythrin from Gloeobacter violaceus | | Descriptor: | PHYCOERYTHRIN ALPHA CHAIN, PHYCOERYTHRIN BETA SUBUNIT, PHYCOERYTHROBILIN, ... | | Authors: | Murray, J.W, Benson, S, Nield, J, Barber, J. | | Deposit date: | 2007-12-10 | | Release date: | 2008-03-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structures of the Phycobiliproteins of Gloeobacter Violaceus
To be Published
|
|
2V8A
 
 | | The structure of Thermosynechococcus elongatus allophycocyanin at 3.5 Angstroems. | | Descriptor: | ALLOPHYCOCYANIN ALPHA CHAIN, ALLOPHYCOCYANIN BETA CHAIN, PHYCOCYANOBILIN | | Authors: | Murray, J.W, Maghlaoui, K, Barber, J. | | Deposit date: | 2007-08-06 | | Release date: | 2007-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The Structure of Allophycocyanin from Thermosynechococcus Elongatus at 3.5 A Resolution.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2VML
 
 | |
2VJR
 
 | |
6RK0
 
 | | Structure of the Flavocytochrome Anf3 from Azotobacter vinelandii | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Murray, J.W, Varghese, F, Kabasakal, B. | | Deposit date: | 2019-04-29 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | A low-potential terminal oxidase associated with the iron-only nitrogenase from the nitrogen-fixing bacteriumAzotobacter vinelandii.
J.Biol.Chem., 294, 2019
|
|
4YC5
 
 | | Beta1 synthetic solenoid protein | | Descriptor: | CHLORIDE ION, beta1 | | Authors: | Murray, J.W. | | Deposit date: | 2015-02-19 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | Synthetic beta-solenoid proteins with the fragment-free computational design of a beta-hairpin extension.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4YFO
 
 | | beta1_ex1 | | Descriptor: | beta1_ex1 | | Authors: | Murray, J.W, MacDonald, J.T. | | Deposit date: | 2015-02-25 | | Release date: | 2016-03-16 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | Synthetic beta-solenoid proteins with the fragment-free computational design of a beta-hairpin extension.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5MLF
 
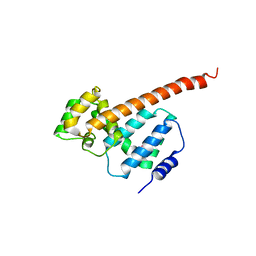 | | Structure of Psb29 at 1.55A | | Descriptor: | MERCURY (II) ION, Protein Thf1 | | Authors: | Murray, J.W, Kozlo, A. | | Deposit date: | 2016-12-06 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.637 Å) | | Cite: | Structure of Psb29/Thf1 and its association with the FtsH protease complex involved in photosystem II repair in cyanobacteria.
Philos. Trans. R. Soc. Lond., B, Biol. Sci., 372, 2017
|
|
4YDT
 
 | | Beta1 synthetic solenoid protein | | Descriptor: | beta1 protein | | Authors: | Murray, J.W. | | Deposit date: | 2015-02-23 | | Release date: | 2016-03-09 | | Last modified: | 2016-10-12 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Synthetic beta-solenoid proteins with the fragment-free computational design of a beta-hairpin extension.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4YCQ
 
 | | Beta1 synthetic solenoid protein | | Descriptor: | beta1 protein | | Authors: | Murray, J.W. | | Deposit date: | 2015-02-20 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Synthetic beta-solenoid proteins with the fragment-free computational design of a beta-hairpin extension.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4YEI
 
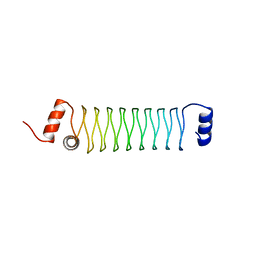 | | Beta1mut synthetic solenoid protein | | Descriptor: | beta1mut | | Authors: | Murray, J.W, Kraatz, S. | | Deposit date: | 2015-02-24 | | Release date: | 2016-03-09 | | Last modified: | 2016-10-12 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Synthetic beta-solenoid proteins with the fragment-free computational design of a beta-hairpin extension.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5MJR
 
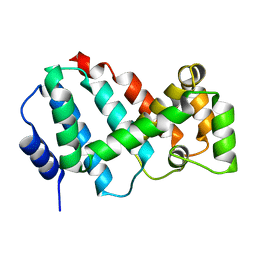 | | Structure of Psb29 at 1.55A | | Descriptor: | Protein Thf1, SULFATE ION | | Authors: | Murray, J.W, Kozlo, A. | | Deposit date: | 2016-12-01 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structure of Psb29/Thf1 and its association with the FtsH protease complex involved in photosystem II repair in cyanobacteria.
Philos. Trans. R. Soc. Lond., B, Biol. Sci., 372, 2017
|
|
5MJO
 
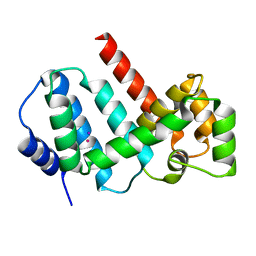 | | Structure of Psb29 at 1.55A | | Descriptor: | IODIDE ION, MERCURY (II) ION, Protein Thf1 | | Authors: | Murray, J.W, Kozlo, A. | | Deposit date: | 2016-12-01 | | Release date: | 2017-08-23 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of Psb29/Thf1 and its association with the FtsH protease complex involved in photosystem II repair in cyanobacteria.
Philos. Trans. R. Soc. Lond., B, Biol. Sci., 372, 2017
|
|
5MJW
 
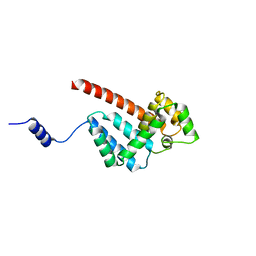 | | Structure of Psb29 at 1.55A | | Descriptor: | Protein Thf1 | | Authors: | Murray, J.W, Kozlo, A. | | Deposit date: | 2016-12-02 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structure of Psb29/Thf1 and its association with the FtsH protease complex involved in photosystem II repair in cyanobacteria.
Philos. Trans. R. Soc. Lond., B, Biol. Sci., 372, 2017
|
|
2C1F
 
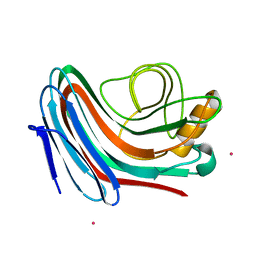 | |
2VA0
 
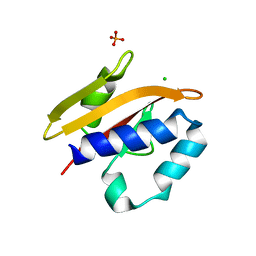 | | Differential regulation of the xylan degrading apparatus of Cellvibrio japonicus by a novel two component system | | Descriptor: | ABFS ARABINOFURANOSIDASE TWO COMPONENT SYSTEM SENSOR PROTEIN, CHLORIDE ION, PHOSPHATE ION | | Authors: | Murray, J.W, Emami, K, Topakas, E, Nagy, T, Henshaw, J, Jackson, K.A, Nelson, K.E, Mongodin, E.F, Lewis, R.J, Gilbert, H.J. | | Deposit date: | 2007-08-28 | | Release date: | 2008-10-14 | | Last modified: | 2011-09-28 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Regulation of the Xylan-Degrading Apparatus of Cellvibrio Japonicus by a Novel Two-Component System.
J.Biol.Chem., 284, 2009
|
|
5SVB
 
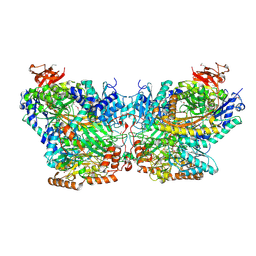 | | Mechanism of ATP-Dependent Acetone Carboxylation, Acetone Carboxylase AMP bound structure | | Descriptor: | ADENOSINE MONOPHOSPHATE, Acetone carboxylase alpha subunit, Acetone carboxylase beta subunit, ... | | Authors: | Eilers, B.J, Mus, F, Alleman, A.B, Kabasakal, B.V, Murray, J.W, Nocek, B.P, Dubois, J.L, Peters, J.W. | | Deposit date: | 2016-08-05 | | Release date: | 2017-08-09 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.645 Å) | | Cite: | Structural Basis for the Mechanism of ATP-Dependent Acetone Carboxylation.
Sci Rep, 7, 2017
|
|
5SVC
 
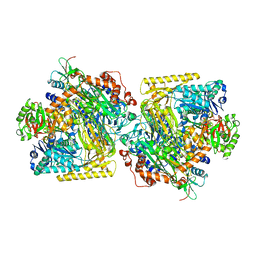 | | Mechanism of ATP-Dependent Acetone Carboxylation, Acetone Carboxylase nucleotide-free structure | | Descriptor: | Acetone carboxylase alpha subunit, Acetone carboxylase beta subunit, Acetone carboxylase gamma subunit, ... | | Authors: | Eilers, B.J, Mus, F, Alleman, A.B, Kabasakal, B.V, Murray, J.W, Nocek, B.P, Dubois, J.L, Peters, J.W. | | Deposit date: | 2016-08-05 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for the Mechanism of ATP-Dependent Acetone Carboxylation.
Sci Rep, 7, 2017
|
|
8AEQ
 
 | |
8AEO
 
 | |
