8GUH
 
 | | Serine Palmitoyltransferase from Sphingobacterium multivorum complexed with Tris | | Descriptor: | 1,2-ETHANEDIOL, Serine palmitoyltransferase, [4-[[[2-(hydroxymethyl)-1,3-bis(oxidanyl)propan-2-yl]amino]methyl]-6-methyl-5-oxidanyl-pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Murakami, T, Takahashi, A, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2022-09-12 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of Sphingobacterium multivorum serine palmitoyltransferase complexed with tris(hydroxymethyl)aminomethane.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
8H29
 
 | | Serine Palmitoyltransferase from Sphingobacterium multivorum complexed with L-threonine | | Descriptor: | 1,2-ETHANEDIOL, N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-L-threonine, Serine palmitoyltransferase | | Authors: | Murakami, T, Takahashi, A, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2022-10-05 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural insights into the substrate recognition of serine palmitoyltransferase from Sphingobacterium multivorum.
J.Biol.Chem., 299, 2023
|
|
8H1Q
 
 | | Serine Palmitoyltransferase from Sphingobacterium multivorum complexed with L-serine | | Descriptor: | 1,2-ETHANEDIOL, Serine palmitoyltransferase, [3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-SERINE | | Authors: | Murakami, T, Takahashi, A, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2022-10-03 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into the substrate recognition of serine palmitoyltransferase from Sphingobacterium multivorum.
J.Biol.Chem., 299, 2023
|
|
8H21
 
 | | Serine Palmitoyltransferase from Sphingobacterium multivorum complexed with L-alanine | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-PROPIONIC ACID, Serine palmitoyltransferase | | Authors: | Murakami, T, Takahashi, A, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2022-10-04 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural insights into the substrate recognition of serine palmitoyltransferase from Sphingobacterium multivorum.
J.Biol.Chem., 299, 2023
|
|
8H1Y
 
 | | Serine Palmitoyltransferase from Sphingobacterium multivorum complexed with L-homoserine | | Descriptor: | (2~{S})-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]-4-oxidanyl-butanoic acid, 1,2-ETHANEDIOL, Serine palmitoyltransferase | | Authors: | Murakami, T, Takahashi, A, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2022-10-04 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insights into the substrate recognition of serine palmitoyltransferase from Sphingobacterium multivorum.
J.Biol.Chem., 299, 2023
|
|
8H20
 
 | | Serine Palmitoyltransferase from Sphingobacterium multivorum complexed with Glycine | | Descriptor: | 1,2-ETHANEDIOL, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], Serine palmitoyltransferase | | Authors: | Murakami, T, Takahashi, A, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2022-10-04 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural insights into the substrate recognition of serine palmitoyltransferase from Sphingobacterium multivorum.
J.Biol.Chem., 299, 2023
|
|
8IYT
 
 | | Crystal Structure of Serine Palmitoyltransferase complexed with D-methylserine | | Descriptor: | (2~{R})-2-methyl-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]-3-oxidanyl-propanoic acid, 1,2-ETHANEDIOL, Serine palmitoyltransferase | | Authors: | Takahashi, A, Murakami, T, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2023-04-06 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Serine Palmitoyltransferase from Sphingobacterium multivorum
To Be Published
|
|
8IYP
 
 | | Crystal structure of serine palmitoyltransferase soaked in 190 mM D-serine solution | | Descriptor: | 1,2-ETHANEDIOL, Serine palmitoyltransferase, [3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-SERINE | | Authors: | Takahashi, A, Murakami, T, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2023-04-05 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Crystal structure of Serine Palmitoyltransferase from Sphingobacterium multivorum
To Be Published
|
|
1F2D
 
 | | 1-AMINOCYCLOPROPANE-1-CARBOXYLATE DEAMINASE | | Descriptor: | 1-AMINOCYCLOPROPANE-1-CARBOXYLATE DEAMINASE, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Yao, M, Ose, T, Sugimoto, H, Horiuchi, A, Nakagawa, A, Yokoi, D, Murakami, T, Honma, M, Wakatsuki, S, Tanaka, I. | | Deposit date: | 2000-05-24 | | Release date: | 2000-12-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of 1-aminocyclopropane-1-carboxylate deaminase from Hansenula saturnus.
J.Biol.Chem., 275, 2000
|
|
8H1W
 
 | | Serine Palmitoyltransferase from Sphingobacterium multivorum | | Descriptor: | 1,2-ETHANEDIOL, Serine palmitoyltransferase | | Authors: | Takahashi, A, Murakami, T, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2022-10-04 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insights into the substrate recognition of serine palmitoyltransferase from Sphingobacterium multivorum.
J.Biol.Chem., 299, 2023
|
|
3G7C
 
 | | Structure of the Phosphorylation Mimetic of Occludin C-term Tail | | Descriptor: | Occludin | | Authors: | Tash, B.R, Sundstrom, J.M, Murakami, T, Flanagan, J.M, Bewley, M.C, Antonetii, D.A. | | Deposit date: | 2009-02-09 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification and analysis of occludin phosphosites: a combined mass spectrometry and bioinformatics approach.
J.PROTEOME RES., 8, 2009
|
|
5B56
 
 | | Crystal structure of HIV-1 VPR C-Terminal domain and DIBB-M-Importin-Alpha2 complex | | Descriptor: | Importin subunit alpha-1, Protein Vpr | | Authors: | Miyatake, H, Sanjoh, A, Matusda, G, Murakami, T, Murakami, H, Hagiwara, K, Yokoyama, M, Sato, H, Miyamoto, Y, Dohmae, N, Aida, Y. | | Deposit date: | 2016-04-25 | | Release date: | 2016-06-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Mechanism of HIV-1 Vpr for Binding to Importin-alpha
J.Mol.Biol., 428, 2016
|
|
3O7R
 
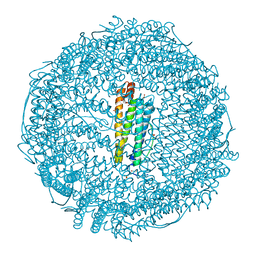 | | Crystal structure of Ru(p-cymene)/apo-H49AFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, Ferritin light chain, ... | | Authors: | Takezawa, Y, Bockmann, P, Sugi, N, Wang, Z, Abe, S, Murakami, T, Hikage, T, Erker, G, Watanabe, Y, Kitagawa, S, Ueno, T. | | Deposit date: | 2010-07-31 | | Release date: | 2011-04-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Incorporation of organometallic Ru complexes into apo-ferritin cage.
J.CHEM.SOC.,DALTON TRANS., 40, 2011
|
|
3O7S
 
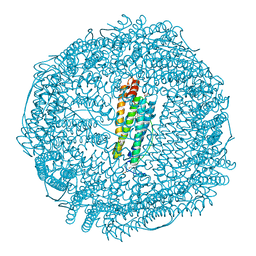 | | Crystal structure of Ru(p-cymene)/apo-Fr | | Descriptor: | 1,2-ETHANEDIOL, 1-methyl-4-(1-methylethyl)benzene, CADMIUM ION, ... | | Authors: | Takezawa, Y, Bockmann, P, Sugi, N, Wang, Z, Abe, S, Murakami, T, Hikage, T, Erker, G, Watanabe, Y, Kitagawa, S, Ueno, T. | | Deposit date: | 2010-07-31 | | Release date: | 2011-04-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Incorporation of organometallic Ru complexes into apo-ferritin cage.
J.Chem.Soc.,Dalton Trans., 40, 2011
|
|
6KGB
 
 | |
1AXJ
 
 | |
7P6T
 
 | | Crystal structure of the FimH-binding decoy module of human glycoprotein 2 (GP2) (crystal form III) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-ethyl-2-(hydroxymethyl)propane-1,3-diol, Isoform Alpha of Pancreatic secretory granule membrane major glycoprotein GP2 | | Authors: | Stsiapanava, A, Tunyasuvunakool, K, Jumper, J, de Sanctis, D, Jovine, L. | | Deposit date: | 2021-07-17 | | Release date: | 2022-03-16 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7P6R
 
 | | Crystal structure of the FimH-binding decoy module of human glycoprotein 2 (GP2) (crystal form I) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform Alpha of Pancreatic secretory granule membrane major glycoprotein GP2 | | Authors: | Stsiapanava, A, Tunyasuvunakool, K, Jumper, J, de Sanctis, D, Jovine, L. | | Deposit date: | 2021-07-17 | | Release date: | 2022-03-16 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7P6S
 
 | | Crystal structure of the FimH-binding decoy module of human glycoprotein 2 (GP2) (crystal form II) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform Alpha of Pancreatic secretory granule membrane major glycoprotein GP2, pentane-1,5-diol | | Authors: | Stsiapanava, A, Tunyasuvunakool, K, Jumper, J, de Sanctis, D, Jovine, L. | | Deposit date: | 2021-07-17 | | Release date: | 2022-03-16 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
1J0D
 
 | | ACC deaminase mutant complexed with ACC | | Descriptor: | 1-aminocyclopropane-1-carboxylate deaminase, N-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-Y-LMETHYL]-1-AMINO-CYCLOPROPANECARBOXYLIC ACID | | Authors: | Ose, T, Fujino, A, Yao, M, Honma, M, Tanaka, I. | | Deposit date: | 2002-11-12 | | Release date: | 2003-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reaction intermediate structures of 1-aminocyclopropane-1-carboxylate deaminase: insight into PLP-dependent cyclopropane ring-opening reaction
J.BIOL.CHEM., 278, 2003
|
|
1J0E
 
 | | ACC deaminase mutant reacton intermediate | | Descriptor: | 1-AMINOCYCLOPROPANECARBOXYLIC ACID, 1-aminocyclopropane-1-carboxylate deaminase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ose, T, Fujino, A, Yao, M, Honma, M, Tanaka, I. | | Deposit date: | 2002-11-12 | | Release date: | 2003-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Reaction intermediate structures of 1-aminocyclopropane-1-carboxylate deaminase: insight into PLP-dependent cyclopropane ring-opening reaction
J.BIOL.CHEM., 278, 2003
|
|
3NTC
 
 | | Crystal structure of KD-247 Fab, an anti-V3 antibody that inhibits HIV-1 Entry | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Fab heavy chain, ... | | Authors: | Sarafianos, S.G, Kirby, K.A. | | Deposit date: | 2010-07-03 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis of clade-specific HIV-1 neutralization by humanized anti-V3 monoclonal antibody KD-247.
Faseb J., 29, 2015
|
|
