1IWG
 
 | |
5ZWL
 
 | |
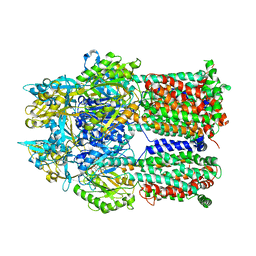
2DRD
 
 | | Crystal structure of a multidrug transporter reveal a functionally rotating mechanism | | Descriptor: | (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, ACRB | | Authors: | Murakami, S, Nakashima, R, Yamashita, E, Matsumoto, T. | | Deposit date: | 2006-06-08 | | Release date: | 2006-08-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of a multidrug transporter reveal a functionally rotating mechanism
Nature, 443, 2006
|
|
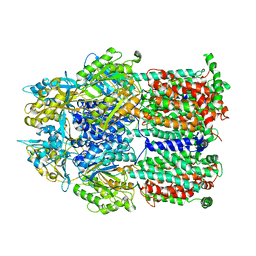
2DR6
 
 | | Crystal structure of a multidrug transporter reveal a functionally rotating mechanism | | Descriptor: | ACRB, DOXORUBICIN | | Authors: | Murakami, S, Nakashima, R, Yamashita, E, Matsumoto, T. | | Deposit date: | 2006-06-08 | | Release date: | 2006-08-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structures of a multidrug transporter reveal a functionally rotating mechanism
Nature, 443, 2006
|
|
2DHH
 
 | |
7CZ9
 
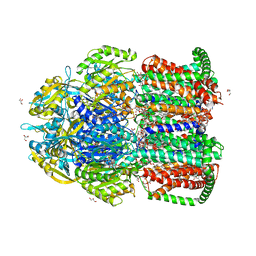 | | Crystal structure of multidrug efflux transporter OqxB from Klebsiella pneumoniae | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Efflux pump membrane transporter, GLYCEROL, ... | | Authors: | Murakami, S, Okada, U, Yamashita, E. | | Deposit date: | 2020-09-07 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and function relationship of OqxB efflux pump from Klebsiella pneumoniae.
Nat Commun, 12, 2021
|
|
5GKO
 
 | |
5WS4
 
 | |
5NIK
 
 | | Structure of the MacAB-TolC ABC-type tripartite multidrug efflux pump | | Descriptor: | Macrolide export ATP-binding/permease protein MacB, Macrolide export protein MacA, Outer membrane protein TolC | | Authors: | Fitzpatrick, A.W.P, Llabres, S, Neuberger, A, Blaza, J.N, Bai, X.-C, Okada, U, Murakami, S, van Veen, H.W, Zachariae, U, Scheres, S.H.W, Luisi, B.F, Du, D. | | Deposit date: | 2017-03-24 | | Release date: | 2017-05-24 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the MacAB-TolC ABC-type tripartite multidrug efflux pump.
Nat Microbiol, 2, 2017
|
|
5NIL
 
 | | Structure of the MacAB-TolC ABC-type tripartite multidrug efflux pump-MacB section | | Descriptor: | Macrolide export ATP-binding/permease protein MacB, Macrolide export protein MacA, Outer membrane protein TolC | | Authors: | Fitzpatrick, A.W.P, Llabres, S, Neuberger, A, Blaza, J.N, Bai, X.-C, Okada, U, Murakami, S, van Veen, H.W, Zachariae, U, Scheres, S.H.W, Luisi, B.F, Du, D. | | Deposit date: | 2017-03-24 | | Release date: | 2017-05-24 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Structure of the MacAB-TolC ABC-type tripartite multidrug efflux pump.
Nat Microbiol, 2, 2017
|
|
4TKB
 
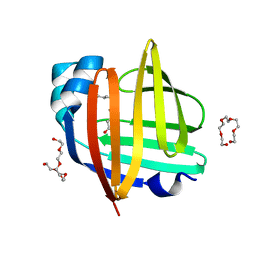 | | The 0.86 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with lauric acid | | Descriptor: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Matsuoka, S, Mizohata, E, Matsuoka, D, Ishida, H, Hirose, M, Kakinouchi, K, Hara, T, Murakami, S, Inoue, T, Murata, M. | | Deposit date: | 2014-05-26 | | Release date: | 2015-01-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.86 Å) | | Cite: | Water-mediated recognition of simple alkyl chains by heart-type Fatty-Acid-binding protein
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4TJZ
 
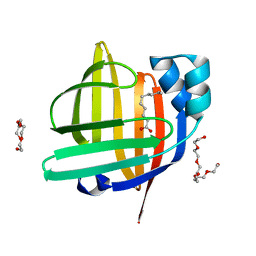 | | The 0.87 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with capric acid | | Descriptor: | DECANOIC ACID, Fatty acid-binding protein, heart, ... | | Authors: | Sugiyama, S, Matsuoka, S, Mizohata, E, Ishida, H, Hirose, M, Kakinouchi, K, Hara, T, Murakami, S, Inoue, T, Murata, M. | | Deposit date: | 2014-05-25 | | Release date: | 2015-01-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.87 Å) | | Cite: | Water-mediated recognition of simple alkyl chains by heart-type Fatty-Acid-binding protein
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4TKH
 
 | | The 0.93 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with myristic acid | | Descriptor: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Matsuoka, S, Mizohata, E, Matsuoka, D, Ishida, H, Hirose, M, Kakinouchi, K, Hara, T, Murakami, S, Inoue, T, Murata, M. | | Deposit date: | 2014-05-26 | | Release date: | 2015-01-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | Water-mediated recognition of simple alkyl chains by heart-type Fatty-Acid-binding protein
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4TKJ
 
 | | The 0.87 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with palmitic acid | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, Fatty acid-binding protein, heart, ... | | Authors: | Sugiyama, S, Matsuoka, S, Mizohata, E, Matsuoka, D, Ishida, H, Hirose, M, Kakinouchi, K, Hara, T, Murakami, S, Inoue, T, Murata, M. | | Deposit date: | 2014-05-26 | | Release date: | 2015-01-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.87 Å) | | Cite: | Water-mediated recognition of simple alkyl chains by heart-type Fatty-Acid-binding protein
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
7WLS
 
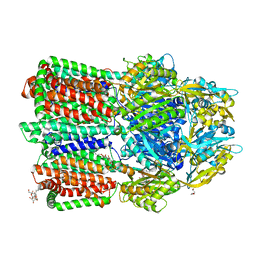 | | Crystal structure of the multidrug efflux transporter BpeB from Burkholderia pseudomallei | | Descriptor: | Efflux pump membrane transporter, TETRAETHYLENE GLYCOL, UNDECYL-MALTOSIDE | | Authors: | Kato, T, Hung, L.-W, Yamashita, E, Okada, U, Terwilliger, T.C, Murakami, S. | | Deposit date: | 2022-01-13 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Crystal structures of multidrug efflux transporters from Burkholderia pseudomallei suggest details of transport mechanism.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7WLV
 
 | | Crystal Structure of the Multidrug effulx transporter BpeF from Burkholderia pseudomallei. | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Efflux pump membrane transporter | | Authors: | Kato, T, Hung, L.-W, Yamashita, E, Okada, U, Terwilliger, T.C, Murakami, S. | | Deposit date: | 2022-01-13 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of multidrug efflux transporters from Burkholderia pseudomallei suggest details of transport mechanism.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3AGV
 
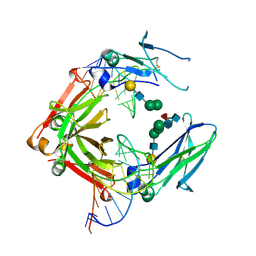 | | Crystal structure of a human IgG-aptamer complex | | Descriptor: | 5'-R(*GP*GP*AP*GP*GP*(UFT)P*GP*(CFZ)P*(UFT)P*(CFZ)P*(CFZ)P*GP*AP*AP*A*GP*GP*AP*AP*(CFZ)P*(UFT)P*(CFZ)P*(CFZ)P*A)-3', CALCIUM ION, Ig gamma-1 chain C region, ... | | Authors: | Nomura, Y, Sugiyama, S, Sakamoto, T, Miyakawa, S, Adachi, H, Takano, K, Murakami, S, Inoue, T, Mori, Y, Nakamura, Y, Matsumura, H. | | Deposit date: | 2010-04-08 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Conformational plasticity of RNA for target recognition as revealed by the 2.15 A crystal structure of a human IgG-aptamer complex
Nucleic Acids Res., 38, 2010
|
|
3WVM
 
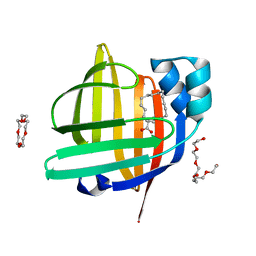 | | The 0.88 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with stearic acid | | Descriptor: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Matsuoka, S, Mizohata, E, Matsuoka, D, Ishida, H, Hirose, M, Kakinouchi, K, Hara, T, Matsumura, H, Murakami, S, Inoue, T, Murata, M. | | Deposit date: | 2014-05-25 | | Release date: | 2015-01-28 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (0.88 Å) | | Cite: | Water-mediated recognition of simple alkyl chains by heart-type fatty-acid-binding protein.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
3WR7
 
 | | Crystal Structure of Spermidine Acetyltransferase from Escherichia coli | | Descriptor: | COENZYME A, SPERMIDINE, Spermidine N1-acetyltransferase | | Authors: | Sugiyama, S, Ishikawa, S, Tomitori, S, Niiyama, M, Hirose, M, Miyazaki, Y, Higashi, K, Adachi, H, Takano, K, Murakami, S, Inoue, T, Mori, Y, Kashiwagi, K, Igarashi, K, Matsumura, H. | | Deposit date: | 2014-02-20 | | Release date: | 2015-09-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism underlying promiscuous polyamine recognition by spermidine acetyltransferase
Int.J.Biochem.Cell Biol., 76, 2016
|
|
2HI7
 
 | | Crystal structure of DsbA-DsbB-ubiquinone complex | | Descriptor: | Disulfide bond formation protein B, Thiol:disulfide interchange protein dsbA, UBIQUINONE-1, ... | | Authors: | Inaba, K, Murakami, S, Suzuki, M, Nakagawa, A, Yamashita, E, Okada, K, Ito, K. | | Deposit date: | 2006-06-29 | | Release date: | 2006-12-05 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal Structure of the DsbB-DsbA Complex Reveals a Mechanism of Disulfide Bond Generation
Cell(Cambridge,Mass.), 127, 2006
|
|
3WBG
 
 | | Structure of the human heart fatty acid-binding protein in complex with 1-anilinonaphtalene-8-sulphonic acid | | Descriptor: | 8-ANILINO-1-NAPHTHALENE SULFONATE, Fatty acid-binding protein, heart | | Authors: | Hirose, M, Sugiyama, S, Ishida, H, Niiyama, M, Matsuoka, D, Hara, T, Sato, F, Mizohata, E, Murakami, S, Inoue, T, Matsuoka, S, Murata, M. | | Deposit date: | 2013-05-16 | | Release date: | 2013-10-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of the human-heart fatty-acid-binding protein 3 in complex with the fluorescent probe 1-anilinonaphthalene-8-sulphonic acid
J.SYNCHROTRON RADIAT., 20, 2013
|
|
8IN9
 
 | | The structure of the GfsA KSQ-AT didomain in complex with the GfsA ACP domain | | Descriptor: | N-[2-(acetylamino)ethyl]-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide, Polyketide synthase | | Authors: | Chisuga, T, Murakami, S, Miyanaga, A, Kudo, F, Eguchi, T. | | Deposit date: | 2023-03-09 | | Release date: | 2023-05-31 | | Last modified: | 2023-06-28 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure-Based Analysis of Transient Interactions between Ketosynthase-like Decarboxylase and Acyl Carrier Protein in a Loading Module of Modular Polyketide Synthase.
Acs Chem.Biol., 18, 2023
|
|
2ZUP
 
 | | Updated crystal structure of DsbB-DsbA complex from E. coli | | Descriptor: | Disulfide bond formation protein B, Thiol:disulfide interchange protein dsbA, UBIQUINONE-1, ... | | Authors: | Inaba, K, Suzuki, M, Murakami, S, Nakagawa, A. | | Deposit date: | 2008-10-28 | | Release date: | 2009-04-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Dynamic nature of disulphide bond formation catalysts revealed by crystal structures of DsbB
Embo J., 28, 2009
|
|
2ZUQ
 
 | | Crystal structure of DsbB-Fab complex | | Descriptor: | Disulfide bond formation protein B, Fab fragment heavy chain, Fab fragment light chain, ... | | Authors: | Inaba, K, Suzuki, M, Murakami, S. | | Deposit date: | 2008-10-28 | | Release date: | 2009-04-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Dynamic nature of disulphide bond formation catalysts revealed by crystal structures of DsbB
Embo J., 28, 2009
|
|
5AVH
 
 | | The 0.90 angstrom structure (I222) of glucose isomerase crystallized in high-strength agarose hydrogel | | Descriptor: | Xylose isomerase | | Authors: | Sugiyama, S, Shimizu, N, Maruyama, N, Sazaki, G, Adachi, H, Takano, K, Murakami, S, Inoue, T, Mori, Y, Matsumura, H. | | Deposit date: | 2015-06-16 | | Release date: | 2015-07-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Growth of protein crystals in hydrogels prevents osmotic shock
J.Am.Chem.Soc., 134, 2012
|
|
