4JSO
 
 | |
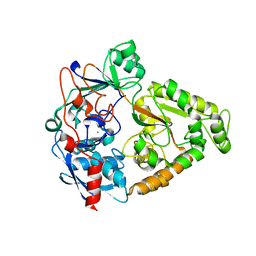
4JSD
 
 | |
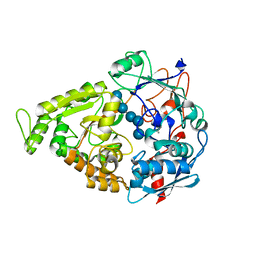
4LNC
 
 | | Neutron structure of the cyclic glucose bound Xylose Isomerase E186Q mutant | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, Xylose isomerase, ... | | Authors: | Munshi, P, Meilleur, F, Myles, D. | | Deposit date: | 2013-07-11 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | NEUTRON DIFFRACTION (2.19 Å) | | Cite: | Neutron structure of the cyclic glucose-bound xylose isomerase E186Q mutant.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3SS2
 
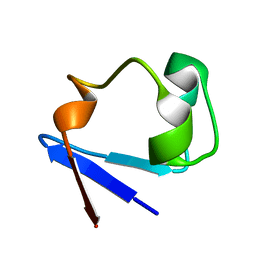 | | Neutron structure of perdeuterated rubredoxin using 48 hours 3rd pass data | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Munshi, P, Chung, C.-L, Blakeley, M.P, Weiss, K.L, Myles, D.A.A, Meilleur, F. | | Deposit date: | 2011-07-07 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | NEUTRON DIFFRACTION (1.75 Å) | | Cite: | Rapid visualization of hydrogen positions in protein neutron crystallographic structures.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3RZ6
 
 | | Neutron structure of perdeuterated rubredoxin using 40 hours 1st pass data | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Munshi, P, Chung, C.-L, Weiss, K.L, Blakeley, M.P, Myles, D.A.A, Meilleur, F. | | Deposit date: | 2011-05-11 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | NEUTRON DIFFRACTION (1.75 Å) | | Cite: | Rapid visualization of hydrogen positions in protein neutron crystallographic structures.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3RZT
 
 | | Neutron structure of perdeuterated rubredoxin using rapid (14 hours) data | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Munshi, P, Chung, C.-L, Weiss, K.L, Blakeley, M.P, Myles, D.A.A, Meilleur, F. | | Deposit date: | 2011-05-12 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | NEUTRON DIFFRACTION (1.7504 Å) | | Cite: | Rapid visualization of hydrogen positions in protein neutron crystallographic structures.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3RYG
 
 | | 128 hours neutron structure of perdeuterated rubredoxin | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Munshi, P, Chung, C.-L, Weiss, K.L, Blakeley, M.P, Myles, D.A.A, Meilleur, F. | | Deposit date: | 2011-05-11 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | NEUTRON DIFFRACTION (1.75 Å) | | Cite: | Rapid visualization of hydrogen positions in protein neutron crystallographic structures.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4K9F
 
 | | Neutron structure of Perdeuterated Rubredoxin refined against 1.75 resolution data collected on the new IMAGINE instrument at HFIR, ORNL | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Munshi, P, Meilleur, F, Myles, D. | | Deposit date: | 2013-04-19 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | NEUTRON DIFFRACTION (1.75 Å) | | Cite: | The IMAGINE instrument: first neutron protein structure and new capabilities for neutron macromolecular crystallography.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
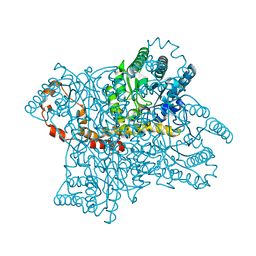
4REK
 
 | | Crystal structure and charge density studies of cholesterol oxidase from Brevibacterium sterolicum at 0.74 ultra-high resolution | | Descriptor: | Cholesterol oxidase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Zarychta, B, Lyubimov, A, Ahmed, M, Munshi, P, Guillot, B, Vrielink, A. | | Deposit date: | 2014-09-23 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.74 Å) | | Cite: | Cholesterol oxidase: ultrahigh-resolution crystal structure and multipolar atom model-based analysis.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
