1BCV
 
 | | SYNTHETIC PEPTIDE CORRESPONDING TO THE MAJOR IMMUNOGEN SITE OF FMD VIRUS, NMR, 10 STRUCTURES | | Descriptor: | PEPTIDE CORRESPONDING TO THE MAJOR IMMUNOGEN SITE OF FMD VIRUS | | Authors: | Petit, M.C, Benkirane, N, Guichard, G, Phan Chan Du, A, Cung, M.T, Briand, J.P, Muller, S. | | Deposit date: | 1998-05-03 | | Release date: | 1998-11-25 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a retro-inverso peptide analogue mimicking the foot-and-mouth disease virus major antigenic site. Structural basis for its antigenic cross-reactivity with the parent peptide.
J.Biol.Chem., 274, 1999
|
|
1BFW
 
 | | RETRO-INVERSO ANALOGUE OF THE G-H LOOP OF VP1 IN FOOT-AND-MOUTH-DISEASE (FMD) VIRUS, NMR, 10 STRUCTURES | | Descriptor: | VP1 PROTEIN | | Authors: | Petit, M.C, Benkirane, N, Guichard, G, Phan Chan Du, A, Cung, M.T, Briand, J.P, Muller, S. | | Deposit date: | 1998-05-22 | | Release date: | 1999-01-13 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a retro-inverso peptide analogue mimicking the foot-and-mouth disease virus major antigenic site. Structural basis for its antigenic cross-reactivity with the parent peptide.
J.Biol.Chem., 274, 1999
|
|
5T4U
 
 | | Crystal structure of the bromodomain of human BRPF1 in complex with a quinolinone ligand | | Descriptor: | 1-METHYLQUINOLIN-2(1H)-ONE, NITRATE ION, Peregrin | | Authors: | Tallant, C, Igoe, N, Bayle, E.D, Nunez-Alonso, G, Newman, J.A, Mathea, S, Savitsky, P, Fedorov, O, Brennan, P.E, Muller, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fish, P, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-08-30 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design of a Biased Potent Small Molecule Inhibitor of the Bromodomain and PHD Finger-Containing (BRPF) Proteins Suitable for Cellular and in Vivo Studies.
J. Med. Chem., 60, 2017
|
|
5T4V
 
 | | Crystal structure of the bromodomain of human BRPF1 in complex with NI-48 ligand | | Descriptor: | 1,2-ETHANEDIOL, 4-cyano-N-(7-methoxy-1,4-dimethyl-2-oxo-1,2-dihydroquinolin-6-yl)benzene-1-sulfonamide, FORMIC ACID, ... | | Authors: | Tallant, C, Igoe, N, Bayle, E.D, Nunez-Alonso, G, Newman, J.A, Mathea, S, Savitsky, P, Fedorov, O, Brennan, P.E, Muller, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fish, P, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-08-30 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Design of a Biased Potent Small Molecule Inhibitor of the Bromodomain and PHD Finger-Containing (BRPF) Proteins Suitable for Cellular and in Vivo Studies.
J. Med. Chem., 60, 2017
|
|
1FF3
 
 | | STRUCTURE OF THE PEPTIDE METHIONINE SULFOXIDE REDUCTASE FROM ESCHERICHIA COLI | | Descriptor: | PEPTIDE METHIONINE SULFOXIDE REDUCTASE, SULFATE ION | | Authors: | Tete-Favier, F, Cobessi, D, Boschi-Muller, S, Azza, S, Branlant, G, Aubry, A. | | Deposit date: | 2000-07-25 | | Release date: | 2000-12-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the Escherichia coli peptide methionine sulphoxide reductase at 1.9 A resolution.
Structure Fold.Des., 8, 2000
|
|
1CVQ
 
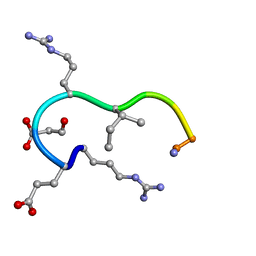 | | SOLUTION STRUCTURE OF THE ANALOGUE RETRO-INVERSO MGREGRIGGC IN CONTACT WITH THE MONOCLONAL ANTIBODY MAB 4X11, NMR, 7 STRUCTURES | | Descriptor: | HISTONE H3 | | Authors: | Phan Chan Du, A, Petit, M.C, Guichard, G, Briand, J.P, Muller, S, Cung, M.T. | | Deposit date: | 1999-08-24 | | Release date: | 1999-09-02 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structure of antibody-bound peptides and retro-inverso analogues. A transferred nuclear Overhauser effect spectroscopy and molecular dynamics approach.
Biochemistry, 40, 2001
|
|
1CWZ
 
 | | Solution structure of the analogue retro-inverso (MA-S)REGRIGGC in contact with the monoclonal antibody MAB 4X11, NMR, 7 structures | | Descriptor: | HISTONE H3, METHYLMALONIC ACID | | Authors: | Phan Chan Du, A, Petit, M.C, Guichard, G, Briand, J.P, Muller, S, Cung, M.T. | | Deposit date: | 1999-08-27 | | Release date: | 1999-09-03 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structure of antibody-bound peptides and retro-inverso analogues. A transferred nuclear Overhauser effect spectroscopy and molecular dynamics approach.
Biochemistry, 40, 2001
|
|
1CT6
 
 | | SOLUTION STRUCTURE OF CGGIRGERG IN CONTACT WITH THE MONOCLONAL ANTIBODY MAB 4X11, NMR, 11 STRUCTURES | | Descriptor: | HISTONE H3 PEPTIDE | | Authors: | Phan Chan Du, A, Petit, M.C, Guichard, G, Briand, J.P, Muller, S, Cung, M.T. | | Deposit date: | 1999-08-19 | | Release date: | 1999-09-02 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structure of antibody-bound peptides and retro-inverso analogues. A transferred nuclear Overhauser effect spectroscopy and molecular dynamics approach.
Biochemistry, 40, 2001
|
|
1CS9
 
 | | SOLUTION STRUCTURE OF CGGIRGERA IN CONTACT WITH THE MONOCLONAL ANTIBODY MAB 4X11, NMR, 7 STRUCTURES | | Descriptor: | HISTONE H3 PEPTIDE | | Authors: | Phan Chan Du, A, Petit, M.C, Guichard, G, Briand, J.P, Muller, S, Cung, M.T. | | Deposit date: | 1999-08-18 | | Release date: | 1999-09-02 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structure of antibody-bound peptides and retro-inverso analogues. A transferred nuclear Overhauser effect spectroscopy and molecular dynamics approach.
Biochemistry, 40, 2001
|
|
1CW8
 
 | | SOLUTION STRUCTURE OF THE ANALOGUE RETRO-INVERSO (mA-R)REGRIGGC IN CONTACT WITH THE MONOCLONAL ANTIBODY MAB 4X11, NMR, 6 STRUCTURES | | Descriptor: | HISTONE H3, METHYLMALONIC ACID | | Authors: | Phan Chan Du, A, Petit, M.C, Guichard, G, Briand, J.P, Muller, S, Cung, T. | | Deposit date: | 1999-08-26 | | Release date: | 1999-09-03 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structure of antibody-bound peptides and retro-inverso analogues. A transferred nuclear Overhauser effect spectroscopy and molecular dynamics approach.
Biochemistry, 40, 2001
|
|
2IEM
 
 | | Solution structure of an oxidized form (Cys51-Cys198) of E. coli Methionine Sulfoxide Reductase A (MsrA) | | Descriptor: | Peptide methionine sulfoxide reductase msrA | | Authors: | Coudevylle, N, Antoine, M, Bouguet-Bonnet, S, Mutzenhardt, P, Boschi-Muller, S, Branlant, G, Cung, M.T. | | Deposit date: | 2006-09-19 | | Release date: | 2007-02-13 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Backbone Dynamics of the Reduced Form and an Oxidized Form of E. coli Methionine Sulfoxide Reductase A (MsrA): Structural Insight of the MsrA Catalytic Cycle.
J.Mol.Biol., 366, 2007
|
|
2FY6
 
 | | Structure of the N-terminal domain of Neisseria meningitidis PilB | | Descriptor: | CHLORIDE ION, Peptide methionine sulfoxide reductase msrA/msrB, SULFATE ION | | Authors: | Ranaivoson, F.M, Kauffmann, B, Neiers, F, Boschi-Muller, S, Branlant, G, Favier, F. | | Deposit date: | 2006-02-07 | | Release date: | 2006-04-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The X-ray Structure of the N-terminal Domain of PILB from Neisseria meningitidis Reveals a Thioredoxin-fold
J.Mol.Biol., 358, 2006
|
|
5MYG
 
 | | Crystal structure of the bromodomain of human BRPF1 in complex with NI-57 chemical probe | | Descriptor: | 4-cyano-~{N}-(1,3-dimethyl-2-oxidanylidene-quinolin-6-yl)-2-methoxy-benzenesulfonamide, Peregrin | | Authors: | Tallant, C, Igoe, N, Bayle, E.D, Krojer, T, Nunez-Alonso, G, Kopec, J, Fitzpatrick, F, Savitsky, P, Fedorov, O, Brennan, P.E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Muller, S, Fish, P, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-01-26 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design of a Chemical Probe for the Bromodomain and Plant Homeodomain Finger-Containing (BRPF) Family of Proteins.
J. Med. Chem., 60, 2017
|
|
5MG2
 
 | | Crystal structure of the second bromodomain of human TAF1 in complex with BAY-299 chemical probe | | Descriptor: | 1,2-ETHANEDIOL, 6-(3-oxidanylpropyl)-2-(1,3,6-trimethyl-2-oxidanylidene-benzimidazol-5-yl)benzo[de]isoquinoline-1,3-dione, Transcription initiation factor TFIID subunit 1 | | Authors: | Tallant, C, Bouche, L, Holton, S.J, Fedorov, O, Siejka, P, Picaud, S, Krojer, T, Srikannathasan, V, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hartung, I.V, Haendler, B, Muller, S, Huber, K.V.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-11-20 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Benzoisoquinolinediones as Potent and Selective Inhibitors of BRPF2 and TAF1/TAF1L Bromodomains.
J. Med. Chem., 60, 2017
|
|
1NQO
 
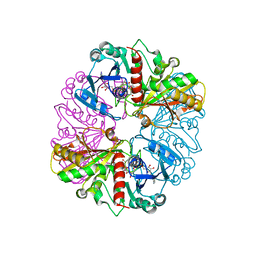 | | Glyceraldehyde-3-Phosphate Dehydrogenase Mutant With Cys 149 Replaced By Ser Complexed With Nad+ and D-Glyceraldehyde-3-Phosphate | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE, Glyceraldehyde 3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Didierjean, C, Corbier, C, Fatih, M, Favier, F, Boschi-Muller, S, Branlant, G, Aubry, A. | | Deposit date: | 2003-01-22 | | Release date: | 2003-04-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of two ternary complexes of phosphorylating Glyceraldehyde-3-Phosphate
Dehydrogenase from Bacillus stearothermophilus with NAD and
D-Glyceraldehyde-3-Phosphate
J.Biol.Chem., 278, 2003
|
|
1NQ5
 
 | | Glyceraldehyde-3-Phosphate Dehydrogenase Mutant With Cys 149 Replaced By Ser Complexed With Nad+ | | Descriptor: | Glyceraldehyde 3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Didierjean, C, Corbier, C, Fatih, M, Favier, F, Boschi-Muller, S, Branlant, G, Aubry, A. | | Deposit date: | 2003-01-21 | | Release date: | 2003-04-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of two ternary complexes of phosphorylating Glyceraldehyde-3-Phosphate Dehydrogenase from Bacillus stearothermophilus with NAD and D-Glyceraldehyde-3-Phosphate
J.Biol.Chem., 278, 2003
|
|
1NPT
 
 | | Glyceraldehyde-3-Phosphate Dehydrogenase Mutant With Cys 149 replaced by Ala complexed with NAD+ | | Descriptor: | Glyceraldehyde 3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Didierjean, C, Corbier, C, Fatih, M, Favier, F, Boschi-Muller, S, Branlant, G, Aubry, A. | | Deposit date: | 2003-01-20 | | Release date: | 2003-04-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal structure of two ternary complexes of phosphorylating Glyceraldehyde-3-Phosphate Dehydrogenase from Bacillus stearothermophilus with NAD and D-Glyceraldehyde-3-Phosphate
J.Biol.Chem., 278, 2003
|
|
1NQA
 
 | | Glyceraldehyde-3-Phosphate Dehydrogenase Mutant With Cys 149 Replaced By Ala Complexed With Nad+ and D-Glyceraldehyde-3-Phosphate | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE, Glyceraldehyde 3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Didierjean, C, Corbier, C, Fatih, M, Favier, F, Boschi-Muller, S, Branlant, G, Aubry, A. | | Deposit date: | 2003-01-21 | | Release date: | 2003-04-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of two ternary complexes of phosphorylating
Glyceraldehyde-3-Phosphate Dehydrogenase from Bacillus stearothermophilus
with NAD and D-Glyceraldehyde-3-Phosphate
J.Biol.Chem., 278, 2003
|
|
5MR8
 
 | | Crystal structure of TRIM33 PHD-Bromodomain isoform B in complex with H3K9ac histone peptide | | Descriptor: | E3 ubiquitin-protein ligase TRIM33, Histone H3, ZINC ION | | Authors: | Tallant, C, Savitsky, P, Fedorov, O, Nunez-Alonso, G, Siejka, P, Krojer, T, Williams, E, Srikannathasan, V, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Muller, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-12-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of TRIM33 PHD-Bromodomain isoform B in complex with H3K9ac histone peptide
To Be Published
|
|
5DKD
 
 | | Crystal structure of the bromodomain of human BRG1 (SMARCA4) in complex with PFI-3 chemical probe | | Descriptor: | (2E)-1-(2-hydroxyphenyl)-3-[(1R,4R)-5-(pyridin-2-yl)-2,5-diazabicyclo[2.2.1]hept-2-yl]prop-2-en-1-one, 1,2-ETHANEDIOL, Transcription activator BRG1, ... | | Authors: | Tallant, C, Owen, D.R, Gerstenberger, B.S, Fedorov, O, Savitsky, P, Nunez-Alonso, G, Newman, J.A, Filippakopoulos, P, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Muller, S, Knapp, S. | | Deposit date: | 2015-09-03 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the bromodomain of human BRG1 (SMARCA4) in complex with PFI-3 chemical probe
To Be Published
|
|
5DKC
 
 | | Crystal structure of the bromodomain of human BRM (SMARCA2) in complex with PFI-3 chemical probe | | Descriptor: | (2E)-1-(2-hydroxyphenyl)-3-[(1R,4R)-5-(pyridin-2-yl)-2,5-diazabicyclo[2.2.1]hept-2-yl]prop-2-en-1-one, Probable global transcription activator SNF2L2, ZINC ION | | Authors: | Tallant, C, Owen, D.R, Gerstenberger, B.S, Fedorov, O, Savitsky, P, Nunez-Alonso, G, Fonseca, M, Krojer, T, Filippakopoulos, P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Muller, S, Knapp, S. | | Deposit date: | 2015-09-03 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the bromodomain of human BRM (SMARCA2) in complex with PFI-3 chemical probe
To Be Published
|
|
5FG6
 
 | | Crystal structure of the bromodomain of human BRD1 (BRPF2) in complex with OF-1 chemical probe | | Descriptor: | 1,2-ETHANEDIOL, 4-bromanyl-~{N}-(6-methoxy-1,3-dimethyl-2-oxidanylidene-benzimidazol-5-yl)-2-methyl-benzenesulfonamide, Bromodomain-containing protein 1, ... | | Authors: | Tallant, C, Owen, D.R, Gerstenberger, B.S, Savitsky, P, Chaikuad, A, Fedorov, O, Nunez-Alonso, G, Filippakopoulos, P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Muller, S, Knapp, S. | | Deposit date: | 2015-12-20 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structure of the bromodomain of human BRD1 (BRPF2) in complex with OF-1 chemical probe
To Be Published
|
|
5FG4
 
 | | Crystal structure of the bromodomain of human BRPF1 in complex with OF-1 chemical probe | | Descriptor: | 4-bromanyl-~{N}-(6-methoxy-1,3-dimethyl-2-oxidanylidene-benzimidazol-5-yl)-2-methyl-benzenesulfonamide, Peregrin | | Authors: | Tallant, C, Owen, D.R, Gerstenberger, B.S, Savitsky, P, Chaikuad, A, Fedorov, O, Nunez-Alonso, G, Filippakopoulos, P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Muller, S, Knapp, S. | | Deposit date: | 2015-12-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the bromodomain of human BRPF1 in complex with OF-1 chemical probe
To Be Published
|
|
5FG5
 
 | | Crystal structure of the bromodomain of human BRPF1 in complex with PFI-4 chemical probe | | Descriptor: | NITRATE ION, Peregrin, ~{N}-(1,3-dimethyl-2-oxidanylidene-6-pyrrolidin-1-yl-benzimidazol-5-yl)-2-methoxy-benzamide | | Authors: | Tallant, C, Owen, D.R, Gerstenberger, B.S, Savitsky, P, Chaikuad, A, Fedorov, O, Nunez-Alonso, G, Filippakopoulos, P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Muller, S, Brennan, P.E, Knapp, S. | | Deposit date: | 2015-12-20 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the bromodomain of human BRPF1 in complex with PFI-4 chemical probe
To Be Published
|
|
5FBX
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with PNZ5 isoxazole inhibitor | | Descriptor: | (3~{S})-5-(3,5-dimethyl-1,2-oxazol-4-yl)-2-methyl-3-phenyl-3~{H}-isoindol-1-one, Bromodomain-containing protein 4 | | Authors: | Tallant, C, Clark, P.G.K, Siejka, P, Fedorov, O, Nowak, R, Srikannathasan, V, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Muller, S, Brennan, P.E, Dixon, D, Knapp, S. | | Deposit date: | 2015-12-14 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the first bromodomain of human BRD4 in complex with PNZ5 isoxazole inhibitor
To Be Published
|
|
