7TAO
 
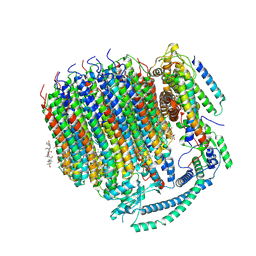 | | Cryo-EM structure of bafilomycin A1 bound to yeast VO V-ATPase | | Descriptor: | (5R)-2,4-dideoxy-1-C-{(2S,3R,4S)-3-hydroxy-4-[(2R,3S,4E,6E,9R,10S,11R,12E,14Z)-10-hydroxy-3,15-dimethoxy-7,9,11,13-tetramethyl-16-oxo-1-oxacyclohexadeca-4,6,12,14-tetraen-2-yl]pentan-2-yl}-4-methyl-5-propan-2-yl-alpha-D-threo-pentopyranose, V-type proton ATPase subunit a, vacuolar isoform, ... | | Authors: | Keon, K.A, Rubinstein, J.L, Benlekbir, S, Kirsch, S.H, Muller, R. | | Deposit date: | 2021-12-21 | | Release date: | 2022-02-23 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM of the Yeast V O Complex Reveals Distinct Binding Sites for Macrolide V-ATPase Inhibitors.
Acs Chem.Biol., 17, 2022
|
|
7TAP
 
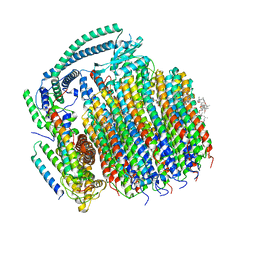 | | Cryo-EM structure of archazolid A bound to yeast VO V-ATPase | | Descriptor: | Archazolid A, V-type proton ATPase subunit a, vacuolar isoform, ... | | Authors: | Keon, K.A, Rubinstein, J.L, Benlekbir, S, Kirsch, S.H, Muller, R. | | Deposit date: | 2021-12-21 | | Release date: | 2022-02-23 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM of the Yeast V O Complex Reveals Distinct Binding Sites for Macrolide V-ATPase Inhibitors.
Acs Chem.Biol., 17, 2022
|
|
4WV3
 
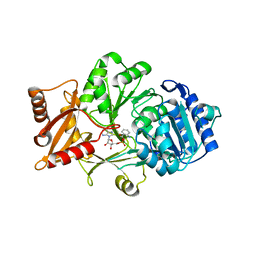 | | Crystal structure of the anthranilate CoA ligase AuaEII in complex with anthranoyl-AMP | | Descriptor: | 5'-O-[(S)-[(2-aminobenzoyl)oxy](hydroxy)phosphoryl]adenosine, Anthranilate-CoA ligase | | Authors: | Tsai, S.C, Muller, R, Jackson, D.R, Tu, S.S, Nguyen, M, Barajas, J.F, Pistorious, D, Krug, D. | | Deposit date: | 2014-11-04 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Structural Insights into Anthranilate Priming during Type II Polyketide Biosynthesis.
Acs Chem.Biol., 11, 2016
|
|
5MIH
 
 | | Crystal structure of the lectin LecA from Pseudomonas aeruginosa in complex with a phenyl-epoxy-galactopyranoside | | Descriptor: | 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CALCIUM ION, PA-I galactophilic lectin, ... | | Authors: | Wagner, S, Hauk, D, Hofmann, M, Joachim, I, Sommer, R, Muller, R, Imberty, A, Varrot, A, Titz, A. | | Deposit date: | 2016-11-28 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Covalent Lectin Inhibition and Application in Bacterial Biofilm Imaging.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
4A10
 
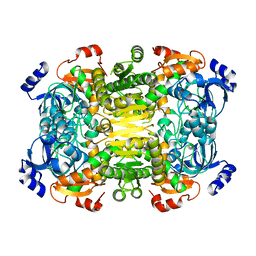 | | Apo-structure of 2-octenoyl-CoA carboxylase reductase CinF from streptomyces sp. | | Descriptor: | OCTENOYL-COA REDUCTASE/CARBOXYLASE | | Authors: | Quade, N, Huo, L, Rachid, S, Heinz, D.W, Muller, R. | | Deposit date: | 2011-09-13 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Unusual Carbon Fixation Giving Rise to Diverse Polyketide Extender Units
Nat.Chem.Biol., 8, 2011
|
|
4A0S
 
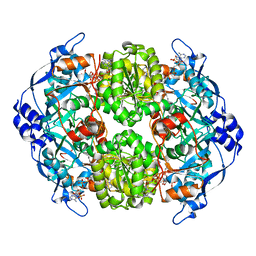 | | STRUCTURE OF THE 2-OCTENOYL-COA CARBOXYLASE REDUCTASE CINF IN COMPLEX WITH NADP AND 2-OCTENOYL-COA | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OCTANOYL-COENZYME A, OCTENOYL-COA REDUCTASE/CARBOXYLASE | | Authors: | Quade, N, Huo, L, Rachid, S, Heinz, D.W, Muller, R. | | Deposit date: | 2011-09-12 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unusual carbon fixation gives rise to diverse polyketide extender units.
Nat. Chem. Biol., 8, 2011
|
|
4JVW
 
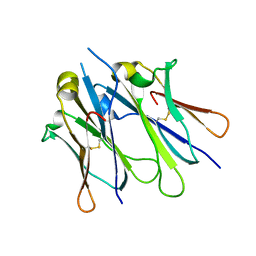 | | IgM C4-domain from mouse | | Descriptor: | Ig mu chain C region secreted form | | Authors: | Mueller, R, Graewert, A.M, Kern, T, Madl, T, Peschek, J, Sattler, M, Groll, M, Buchner, J. | | Deposit date: | 2013-03-26 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structures of the IgM Fc domains reveal principles of its hexamer formation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JVU
 
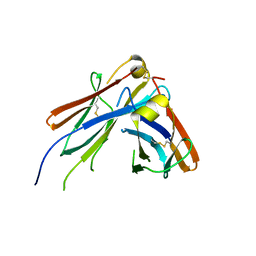 | | IgM C2-domain from mouse | | Descriptor: | Ig mu chain C region membrane-bound form | | Authors: | Mueller, R, Graewert, A.M, Kern, T, Madl, T, Peschek, J, Sattler, M, Groll, M, Buchner, J. | | Deposit date: | 2013-03-26 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High-resolution structures of the IgM Fc domains reveal principles of its hexamer formation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2GJZ
 
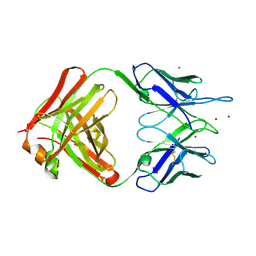 | |
2GK0
 
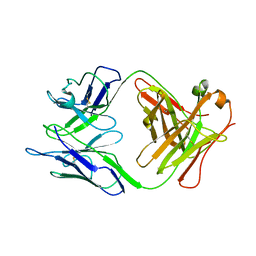 | |
4WID
 
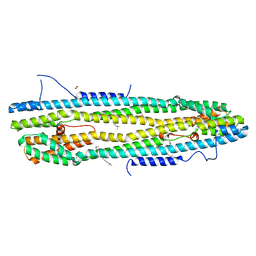 | | Crystal structure of the immediate-early 1 protein (IE1) at 2.31 angstrom (tetragonal form after crystal dehydration) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, RhUL123 | | Authors: | Klingl, S, Scherer, M, Sevvana, M, Muller, Y.A, Stamminger, T. | | Deposit date: | 2014-09-25 | | Release date: | 2014-10-29 | | Last modified: | 2017-09-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal Structure of Cytomegalovirus IE1 Protein Reveals Targeting of TRIM Family Member PML via Coiled-Coil Interactions.
Plos Pathog., 10, 2014
|
|
1BBP
 
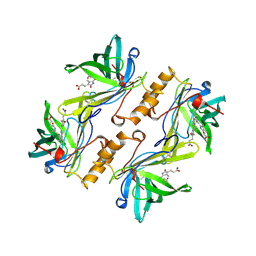 | | MOLECULAR STRUCTURE OF THE BILIN BINDING PROTEIN (BBP) FROM PIERIS BRASSICAE AFTER REFINEMENT AT 2.0 ANGSTROMS RESOLUTION. | | Descriptor: | BILIN BINDING PROTEIN, BILIVERDIN IX GAMMA CHROMOPHORE | | Authors: | Huber, R, Schneider, M, Mayr, I, Mueller, R, Deutzmann, R, Suter, F, Zuber, H, Falk, H, Kayser, H. | | Deposit date: | 1990-09-19 | | Release date: | 1991-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular structure of the bilin binding protein (BBP) from Pieris brassicae after refinement at 2.0 A resolution.
J.Mol.Biol., 198, 1987
|
|
4V2P
 
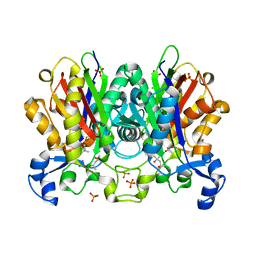 | | Ketosynthase MxnB | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, KETOSYNTHASE, ... | | Authors: | Koehnke, J. | | Deposit date: | 2014-10-13 | | Release date: | 2015-08-26 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | In vitro reconstitution of alpha-pyrone ring formation in myxopyronin biosynthesis.
Chem Sci, 6, 2015
|
|
6VVZ
 
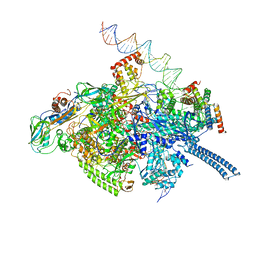 | | Mycobacterium tuberculosis RNAP S456L mutant transcription initiation intermediate structure with Sorangicin | | Descriptor: | DNA (62-MER), DNA (65-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lilic, M, Boyaci, H, Chen, J, Darst, S.A, Campbell, E.A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | The antibiotic sorangicin A inhibits promoter DNA unwinding in a Mycobacterium tuberculosis rifampicin-resistant RNA polymerase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VVT
 
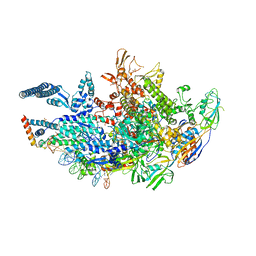 | | Crystal structure of a Mycobacterium smegmatis transcription initiation complex with Rifampicin-resistant RNA polymerase and antibiotic Sorangicin | | Descriptor: | 1,2-ETHANEDIOL, DNA (26-MER), DNA (31-MER), ... | | Authors: | Lilic, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | The antibiotic sorangicin A inhibits promoter DNA unwinding in a Mycobacterium tuberculosis rifampicin-resistant RNA polymerase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VVY
 
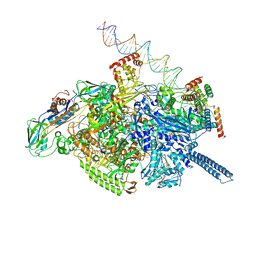 | | Mycobacterium tuberculosis WT RNAP transcription open promoter complex with Sorangicin | | Descriptor: | DNA (65-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Lilic, M, Boyaci, H, Chen, J, Darst, S.A, Campbell, E.A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | The antibiotic sorangicin A inhibits promoter DNA unwinding in a Mycobacterium tuberculosis rifampicin-resistant RNA polymerase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VVV
 
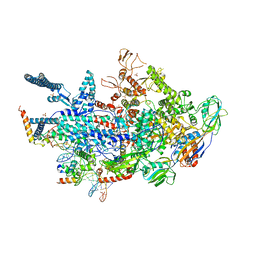 | | Crystal structure of a Mycobacterium smegmatis transcription initiation complex with Rifampicin-resistant RNA polymerase | | Descriptor: | DNA (26-MER), DNA (31-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lilic, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The antibiotic sorangicin A inhibits promoter DNA unwinding in a Mycobacterium tuberculosis rifampicin-resistant RNA polymerase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VW0
 
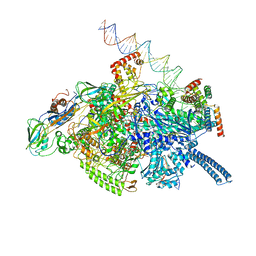 | | Mycobacterium tuberculosis RNAP S456L mutant open promoter complex | | Descriptor: | DNA (65-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Lilic, M, Boyaci, H, Chen, J, Darst, S.A, Campbell, E.A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | The antibiotic sorangicin A inhibits promoter DNA unwinding in a Mycobacterium tuberculosis rifampicin-resistant RNA polymerase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VVS
 
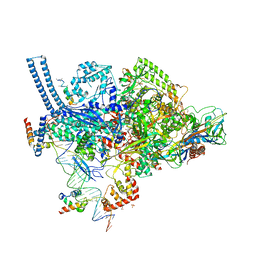 | | Crystal structure of a Mycobacterium smegmatis RNA polymerase transcription initiation complex with antibiotic Sorangicin | | Descriptor: | 1,2-ETHANEDIOL, DNA (26-MER), DNA (31-MER), ... | | Authors: | Lilic, M, Braffman, N, Darst, S.A, Campbell, E.A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.112 Å) | | Cite: | The antibiotic sorangicin A inhibits promoter DNA unwinding in a Mycobacterium tuberculosis rifampicin-resistant RNA polymerase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VVX
 
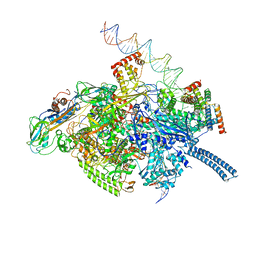 | | Mycobacterium tuberculosis WT RNAP transcription initiation intermediate structure with Sorangicin | | Descriptor: | DNA (63-MER), DNA (65-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lilic, M, Boyaci, H, Chen, J, Campbell, E.A, Darst, S.A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | The antibiotic sorangicin A inhibits promoter DNA unwinding in a Mycobacterium tuberculosis rifampicin-resistant RNA polymerase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8CC4
 
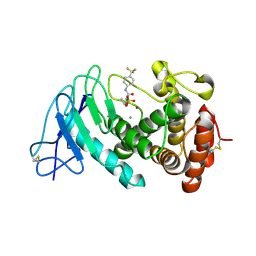 | | LasB bound to phosphonic acid based inhibitor | | Descriptor: | CALCIUM ION, Elastase, ZINC ION, ... | | Authors: | Mueller, R, Sikandar, A. | | Deposit date: | 2023-01-26 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Inhibitors of the Elastase LasB for the Treatment of Pseudomonas aeruginosa Lung Infections.
Acs Cent.Sci., 9, 2023
|
|
7OVR
 
 | | Mature HIV-1 matrix structure | | Descriptor: | HIV-1 matrix, MYRISTIC ACID, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Qu, K, Ke, Z.L, Zila, V, Anders-Oesswein, M, Glass, B, Muecksch, F, Mueller, R, Schultz, C, Mueller, B, Kraeusslich, H.G, Briggs, J.A.G. | | Deposit date: | 2021-06-15 | | Release date: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Maturation of the matrix and viral membrane of HIV-1.
Science, 373, 2021
|
|
7OVQ
 
 | | Immature HIV-1 matrix structure | | Descriptor: | Gag polyprotein, MYRISTIC ACID | | Authors: | Qu, K, Ke, Z.L, Zila, V, Anders-Oesswein, M, Glass, B, Muecksch, F, Mueller, R, Schultz, C, Mueller, B, Kraeusslich, H.G, Briggs, J.A.G. | | Deposit date: | 2021-06-15 | | Release date: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Maturation of the matrix and viral membrane of HIV-1.
Science, 373, 2021
|
|
5LWO
 
 | | Structure of Spin-labelled T4 lysozyme mutant L115C-R119C-R1 at 100K | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Loll, B, Consentius, P, Gohlke, U, Mueller, R, Kaupp, M, Heinemann, U, Wahl, M.C, Risse, T. | | Deposit date: | 2016-09-18 | | Release date: | 2017-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.183 Å) | | Cite: | Internal Dynamics of the 3-Pyrroline-N-Oxide Ring in Spin-Labeled Proteins.
J Phys Chem Lett, 8, 2017
|
|
8OO2
 
 | | ChdA complex with amido-chelocardin | | Descriptor: | 2-carboxamido-2-deacetyl-chelocardin, MAGNESIUM ION, Putative transcriptional regulator | | Authors: | Koehnke, J, Sikandar, A. | | Deposit date: | 2023-04-04 | | Release date: | 2023-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Revision of the Absolute Configurations of Chelocardin and Amidochelocardin.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
