2Z7B
 
 | | Crystal Structure of Mesorhizobium loti 3-hydroxy-2-methylpyridine-4,5-dicarboxylate decarboxylase | | Descriptor: | MANGANESE (II) ION, Mlr6791 protein | | Authors: | McCulloch, K.M, Mukherjee, T, Ealick, S.E, Begley, T.P. | | Deposit date: | 2007-08-17 | | Release date: | 2007-11-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Gene Identification and Structural Characterization of the Pyridoxal 5'-Phosphate Degradative Protein 3-Hydroxy-2-methylpyridine-4,5-dicarboxylate Decarboxylase from Mesorhizobium loti MAFF303099
Biochemistry, 46, 2007
|
|
5WAN
 
 | | Crystal Structure of a flavoenzyme RutA in the pyrimidine catabolic pathway | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Pyrimidine monooxygenase RutA, ... | | Authors: | Zhang, Y, Mukherjee, T, Abdelwahed, S, Begley, T.P, Ealick, S.E. | | Deposit date: | 2017-06-26 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Catalysis of a flavoenzyme-mediated amide hydrolysis.
J. Am. Chem. Soc., 132, 2010
|
|
3R5L
 
 | | Structure of Ddn, the Deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Deazaflavin-dependent nitroreductase | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayyar, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-18 | | Release date: | 2012-01-18 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3R5R
 
 | | Structure of Ddn, the Deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824, with co-factor F420 | | Descriptor: | COENZYME F420, Deazaflavin-dependent nitroreductase | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-19 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3R5Z
 
 | | Structure of a Deazaflavin-dependent reductase from Nocardia farcinica, with co-factor F420 | | Descriptor: | COENZYME F420, Putative uncharacterized protein, SULFATE ION | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-20 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3R5P
 
 | | Structure of Ddn, the Deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824 | | Descriptor: | Deazaflavin-dependent nitroreductase, SULFATE ION | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-19 | | Release date: | 2012-01-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3R5Y
 
 | | Structure of a Deazaflavin-dependent nitroreductase from Nocardia farcinica, with co-factor F420 | | Descriptor: | COENZYME F420, Putative uncharacterized protein | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-20 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3R5W
 
 | | Structure of Ddn, the Deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824, with co-factor F420 | | Descriptor: | COENZYME F420, Deazaflavin-dependent nitroreductase | | Authors: | Cellitti, S.E, Shaffer, J, Jones, D.H, Mukherjee, T, Gurumurthy, M, Bursulaya, B, Boshoff, H.I.M, Choi, I, Nayya, A, Lee, Y.S, Cherian, J, Niyomrattanakit, P, Dick, T, Manjunatha, U.H, Barry, C.E, Spraggon, G, Geierstanger, B.H. | | Deposit date: | 2011-03-20 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.786 Å) | | Cite: | Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824.
Structure, 20, 2012
|
|
3GMB
 
 | | Crystal Structure of 2-Methyl-3-hydroxypyridine-5-carboxylic acid Oxygenase | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | McCulloch, K.M, Mukherjee, T, Begley, T.P, Ealick, S.E. | | Deposit date: | 2009-03-13 | | Release date: | 2009-04-14 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the PLP degradative enzyme 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase from Mesorhizobium loti MAFF303099 and its mechanistic implications.
Biochemistry, 48, 2009
|
|
3GMC
 
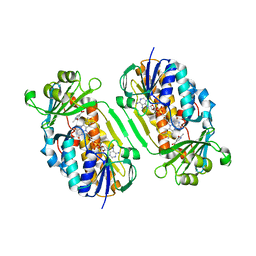 | | Crystal Structure of 2-Methyl-3-hydroxypyridine-5-carboxylic acid Oxygenase with substrate bound | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, 5-hydroxy-6-methylpyridine-3-carboxylic acid, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | McCulloch, K.M, Mukherjee, T, Begley, T.P, Ealick, S.E. | | Deposit date: | 2009-03-13 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the PLP degradative enzyme 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase from Mesorhizobium loti MAFF303099 and its mechanistic implications.
Biochemistry, 48, 2009
|
|
3KXP
 
 | | Crystal Structure of E-2-(Acetamidomethylene)succinate Hydrolase | | Descriptor: | Alpha-(N-acetylaminomethylene)succinic acid hydrolase, CHLORIDE ION | | Authors: | McCulloch, K.M, Mukherjee, T, Begley, T.P, Ealick, S.E. | | Deposit date: | 2009-12-03 | | Release date: | 2010-02-09 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure determination and characterization of the vitamin B(6) degradative enzyme (E)-2-(acetamidomethylene)succinate hydrolase.
Biochemistry, 49, 2010
|
|
2NOX
 
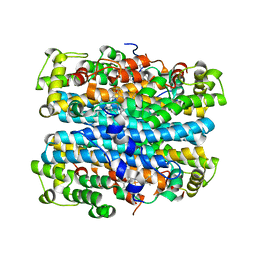 | | Crystal structure of tryptophan 2,3-dioxygenase from Ralstonia metallidurans | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Tryptophan 2,3-dioxygenase | | Authors: | Zhang, Y, Kang, S.A, Mukherjee, T, Bale, S, Crane, B.R, Begley, T.P, Ealick, S.E. | | Deposit date: | 2006-10-26 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and mechanism of tryptophan 2,3-dioxygenase, a heme enzyme involved in tryptophan catabolism and in quinolinate biosynthesis.
Biochemistry, 46, 2007
|
|
